Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00142330Standard Name
RAD51 (Homolog of RADiation sensitive protein family 51)Aliases
PreTt26639 | 11.m00336 | 3702.m00060Description
RAD51 DNA repair RAD51-like protein; DNA repair protein RAD51 containing protein; strand exchange protein; involved in homologous recombination and repair of double-stranded DNA breaks; DNA recombination and repair protein Rad51-like, C-terminalGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- macronucleus (IDA) | GO:0031039
Molecular Function
- ATP-dependent activity, acting on DNA (IDA) | GO:0008094
- double-stranded DNA binding (IDA) | GO:0003690
- single-stranded DNA binding (IDA) | GO:0003697
Biological Process
- DNA damage response (IEP) | GO:0006974
- nuclear division (IMP) | GO:0000280
- regulation of DNA ligation (IDA) | GO:0051105
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
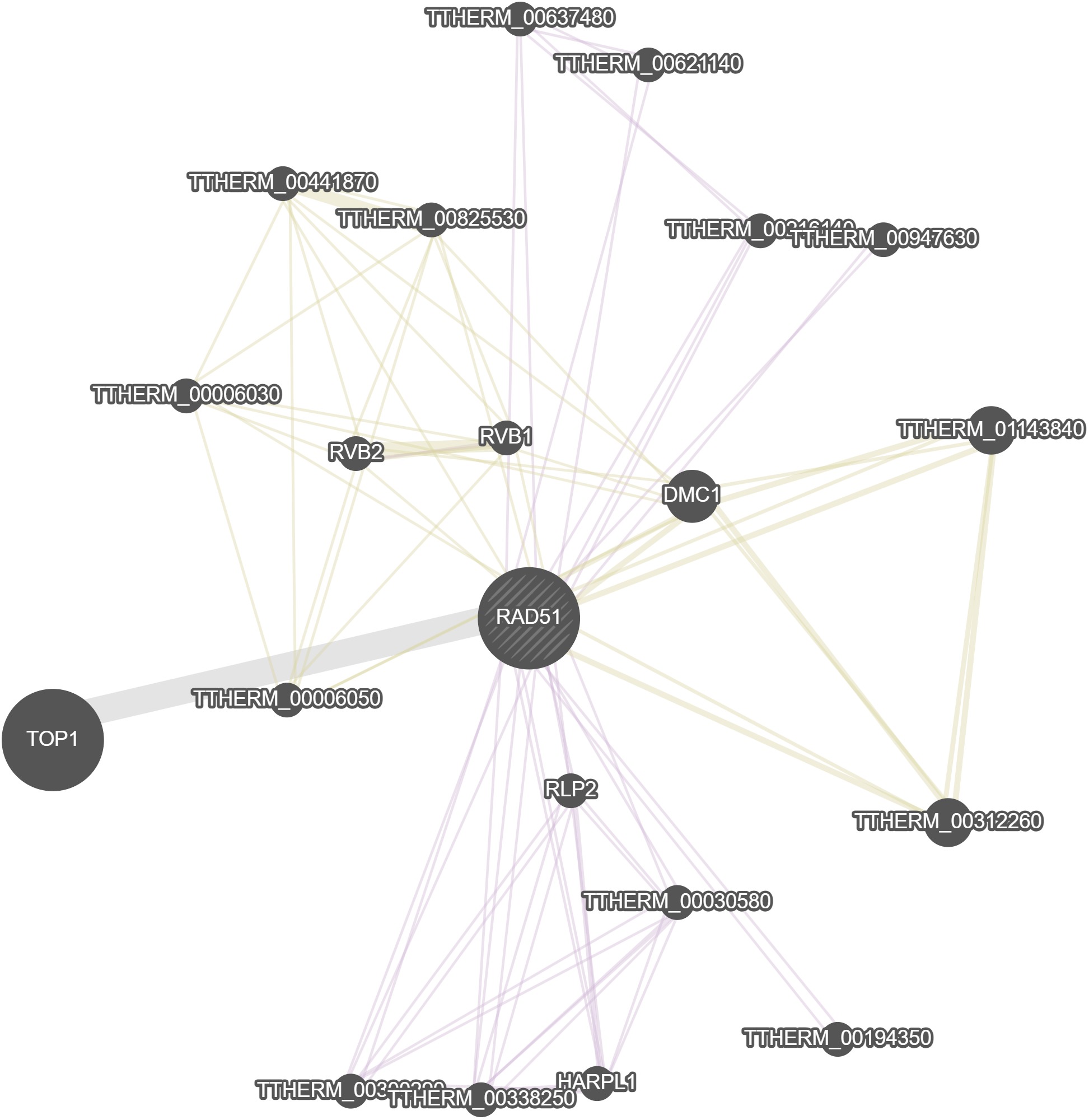
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:34086947: Nabeel-Shah S, Garg J, Saettone A, Ashraf K, Lee H, Wahab S, Ahmed N, Fine J, Derynck J, Pu S, Ponce M, Marcon E, Zhang Z, Greenblatt JF, Pearlman RE, Lambert JP, Fillingham J (2021) Functional characterization of RebL1 highlights the evolutionary conservation of oncogenic activities of the RBBP4/7 orthologue in Tetrahymena thermophila. Nucleic acids research ( ):
- Ref:21483758: Howard-Till RA, Lukaszewicz A, Loidl J (2011) The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. PLoS genetics 7(3):e1001359
- Ref:17005912: Yakisich JS, Sandoval PY, Morrison TL, Kapler GM (2006) TIF1 activates the intra-S-phase checkpoint response in the diploid micronucleus and amitotic polyploid macronucleus of Tetrahymena. Molecular biology of the cell 17(12):5185-97
- Ref:15304567: Smith JJ, Cole ES, Romero DP (2004) Transcriptional control of RAD51 expression in the ciliate Tetrahymena thermophila. Nucleic acids research 32(14):4313-21
- Ref:15522890: Loidl J, Scherthan H (2004) Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. Journal of cell science 117(Pt 24):5791-801
- Ref:11290715: Marsh TC, Cole ES, Romero DP (2001) The transition from conjugal development to the first vegetative cell division is dependent on RAD51 expression in the ciliate Tetrahymena thermophila. Genetics 157(4):1591-8
- Ref:10747055: Marsh TC, Cole ES, Stuart KR, Campbell C, Romero DP (2000) RAD51 is required for propagation of the germinal nucleus in Tetrahymena thermophila. Genetics 154(4):1587-96
- Ref:9628914: Campbell C, Romero DP (1998) Identification and characterization of the RAD51 gene from the ciliate Tetrahymena thermophila. Nucleic acids research 26(13):3165-72
 Sequences
Sequences
>TTHERM_00142330(coding)
ATGGCTGAGTACGCTGAAGACGTTGAATAAAGCGATGGACCTATGTTGATCGCCAAATTG
GAAGAACATGGTATTAATAATGCAGATGTTAAAAAACTCATAGATGCTGGTTTCCAAACT
GTTGAATCCATTTCTTACACTGCTAAGAAAAATCTCTTGCAAATTAAGGGTATGACTGAA
GCAAAAATCGATAAAATCCTTGATGTTGCTGCCAAATTAGTTCCTAATGATTTCTAAACT
GCAGCTGAATACTATGTAAAGAGACAAAGTGTCATCAATCTTACTACAGGCTCAACTGAA
TTAGACAAACTTCTTGGTGGTGGTTTTGAAACAGGCTCTCTCACTGAAATTTTTGGTGAA
TTCAGAACAGGTAAAACTTAAATTTGTCATACTTTGTGTATCACCTGCTAATTACCCAAG
GAAAAGGGTGGTGGTGAAGGTAAGGCTATGTATATTGATACTGAAGGTACTTTCAGACCT
GAAAGACTTGAATCAATTGCTGAAAGATTCGGATTAGATCCTCAAGAATGTATGGAAAAC
GTAGCTTATGCTAGAGCCTTCAACTGTGATCAATAAAATAAACTCTTAGTCCAGGCTGCT
GCTTTGATGGCTGAAAGTAAATACGCTTTGCTTATTGTTGATTCAGCCACTGCTCTTTAC
AGAACTGATTATAGTGGTAGAGGTGAATTGTCAGTCAGACAAAATCACTTGGGTAAATTC
TTGAGAAACTTATAAAGACTTGCTGACGAATTCGGTATTGCTGTAGTTATTACCAACCAA
GTTATGTCCTAAGTCGATGGTGCAGCTATGTTCGCTGGTGATATGAAGAAACCTATTGGT
GGTAATATTATGGCTCACGCTTCTACTACCAGACTTTATCTCAGAAAAGGTAGAGGAGAG
TCTCGTATTTGCAAAATTTACGATTCTCCATGTCTTCCTGAATCAGAAGCTATCTACGCT
ATTGGTAAGGGTGGTATTGAAGACTTCAACGAGTGA>TTHERM_00142330(gene)
AACAAAAACAAATCAAAAAAAAGAATTGCTATATAAGCAAAATTTTAACTTGTTAAATTT
TCTTTAAAATTAATAGTAAATTTATAGACCAAAAATTTAAAAGAAAAAATGGCTGAGTAC
GCTGAAGACGTTGAATAAAGCGATGGACCTATGTTGATCGCCAAATTGGAAGAACATGGT
ATTAATAATGCAGATGTTAAAAAACTCATAGATGCTGGTTTCCAAACTGTTGAATCCATT
TCTTACACTGCTAAGAAAAATCTCTTGCAAATTAAGGGTATGACTGAAGCAAAAATCGAT
AAAATCCTTGATGTTGCTGCCAAATTAGTTCCTAATGATTTCTAAACTGCAGCTGAATAC
TATGTAAAGAGACAAAGTGTCATCAATCTTACTACAGGCTCAACTGAATTAGACAAACTT
CTTGGTGGTGGTTTTGAAACAGGCTCTCTCACTGAAATTTTTGGTGAATTCAGAACAGGT
AAAACTTAAATTTGTCATACTTTGTGTATCACCTGCTAATTACCCAAGGAAAAGGGTGGT
GGTGAAGGTAAGGCTATGTATATTGATACTGAAGGTACTTTCAGACCTGAAAGACTTGAA
TCAATTGCTGAAAGATTCGGATTAGATCCTCAAGAATGTATGGAAAACGTAGCTTATGCT
AGAGCCTTCAACTGTGATCAATAAAATAAACTCTTAGTCCAGGCTGCTGCTTTGATGGCT
GAAAGTAAATACGCTTTGCTTATTGTTGATTCAGCCACTGCTCTTTACAGAACTGATTAT
AGTGGTAGAGGTGAATTGTCAGTCAGACAAAATCACTTGGGTAAATTCTTGAGAAACTTA
TAAAGACTTGCTGACGAATTCGGTATTGCTGTAGTTATTACCAACCAAGTTATGTCCTAA
GTCGATGGTGCAGCTATGTTCGCTGGTGATATGAAGAAACCTATTGGTGGTAATATTATG
GCTCACGCTTCTACTACCAGACTTTATCTCAGAAAAGGTAGAGGAGAGTCTCGTATTTGC
AAAATTTACGATTCTCCATGTCTTCCTGAATCAGAAGCTATCTACGCTATTGGTAAGGGT
GGTATTGAAGACTTCAACGAGTGAAAAAAAAATTTTTAAAGCTAAAAGATAATAAGATAA
AATTCATACTAACTAAATAACTAATAAAATATATAAACTATGCAATTCCAAAATAGATAA
ACAAATATATAAATAGGAGTTATAGAGTATTAATATAATAAAATAATAATTAATTGTACT
CTTTGTTAATCAATCTATATTCTAGCTACTTGTATTTCTTTTACGATATAGATT>TTHERM_00142330(protein)
MAEYAEDVEQSDGPMLIAKLEEHGINNADVKKLIDAGFQTVESISYTAKKNLLQIKGMTE
AKIDKILDVAAKLVPNDFQTAAEYYVKRQSVINLTTGSTELDKLLGGGFETGSLTEIFGE
FRTGKTQICHTLCITCQLPKEKGGGEGKAMYIDTEGTFRPERLESIAERFGLDPQECMEN
VAYARAFNCDQQNKLLVQAAALMAESKYALLIVDSATALYRTDYSGRGELSVRQNHLGKF
LRNLQRLADEFGIAVVITNQVMSQVDGAAMFAGDMKKPIGGNIMAHASTTRLYLRKGRGE
SRICKIYDSPCLPESEAIYAIGKGGIEDFNE