Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00294860Standard Name
CYC33 (CYClin 33)Aliases
PreTt15596 | 27.m00256 | 3692.m00059Description
CYC33 amine-terminal domain cyclin; Cyclin PHO80-likeGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- contractile vacuole (IEA) | GO:0000331
- subrhabdomeral cisterna (IEA) | GO:0016029
Molecular Function
- ferric enterobactin:proton symporter activity (IEA) | GO:0015345
- mercury (II) reductase (NADP+) activity (IEA) | GO:0016152
- obsolete apoptosis activator activity (IEA) | GO:0016506
- obsolete zinc, iron permease activity (IEA) | GO:0015342
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- pyrimidine-nucleoside phosphorylase activity (IEA) | GO:0016154
- siderophore-iron transmembrane transporter activity (IEA) | GO:0015343
Biological Process
- aldosterone biosynthetic process (IEA) | GO:0032342
- bud dilation involved in lung branching (IEA) | GO:0060503
- cichorine biosynthetic process (IEA) | GO:0062032
- ciliary transition fiber assembly (IEA) | GO:1905353
- dorsal closure, leading edge cell fate commitment (IEA) | GO:0035029
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- inferior colliculus development (IEA) | GO:0061379
- lung connective tissue development (IEA) | GO:0060427
- lung epithelium development (IEA) | GO:0060428
- lung saccule development (IEA) | GO:0060430
- memory T cell activation (IEA) | GO:0035709
- mitotic telophase (IEA) | GO:0000093
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- nurse cell nucleus anchoring (IEA) | GO:0007302
- obsolete fertilization, exchange of chromosomal proteins (IEA) | GO:0035042
- obsolete induction by organism of defense-related symbiont reactive oxygen species production (IEA) | GO:0052401
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete positive regulation of Factor XII activation (IEA) | GO:1905160
- obsolete positive regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process (IEA) | GO:1902964
- ommochrome biosynthetic process (IEA) | GO:0006727
- peripheral tolerance induction (IEA) | GO:0002465
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of axon guidance (IEA) | GO:1902669
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of intestinal epithelial cell development (IEA) | GO:1905300
- primary lung bud formation (IEA) | GO:0060431
- protein localization to old growing cell tip (IEA) | GO:1903858
- quercetin metabolic process (IEA) | GO:1901732
- receptor guanylyl cyclase signaling pathway (IEA) | GO:0007168
- regulation of response to oxidative stress (IEA) | GO:1902882
- regulation of stress granule assembly (IEA) | GO:0062028
- somatic recombination of immunoglobulin gene segments (IEA) | GO:0016447
- sperm aster formation (IEA) | GO:0035044
- synaptic vesicle priming (IEA) | GO:0016082
- tRNA import into mitochondrion (IEA) | GO:0016031
 Domains
Domains
- ( PF08613 ) Cyclin
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
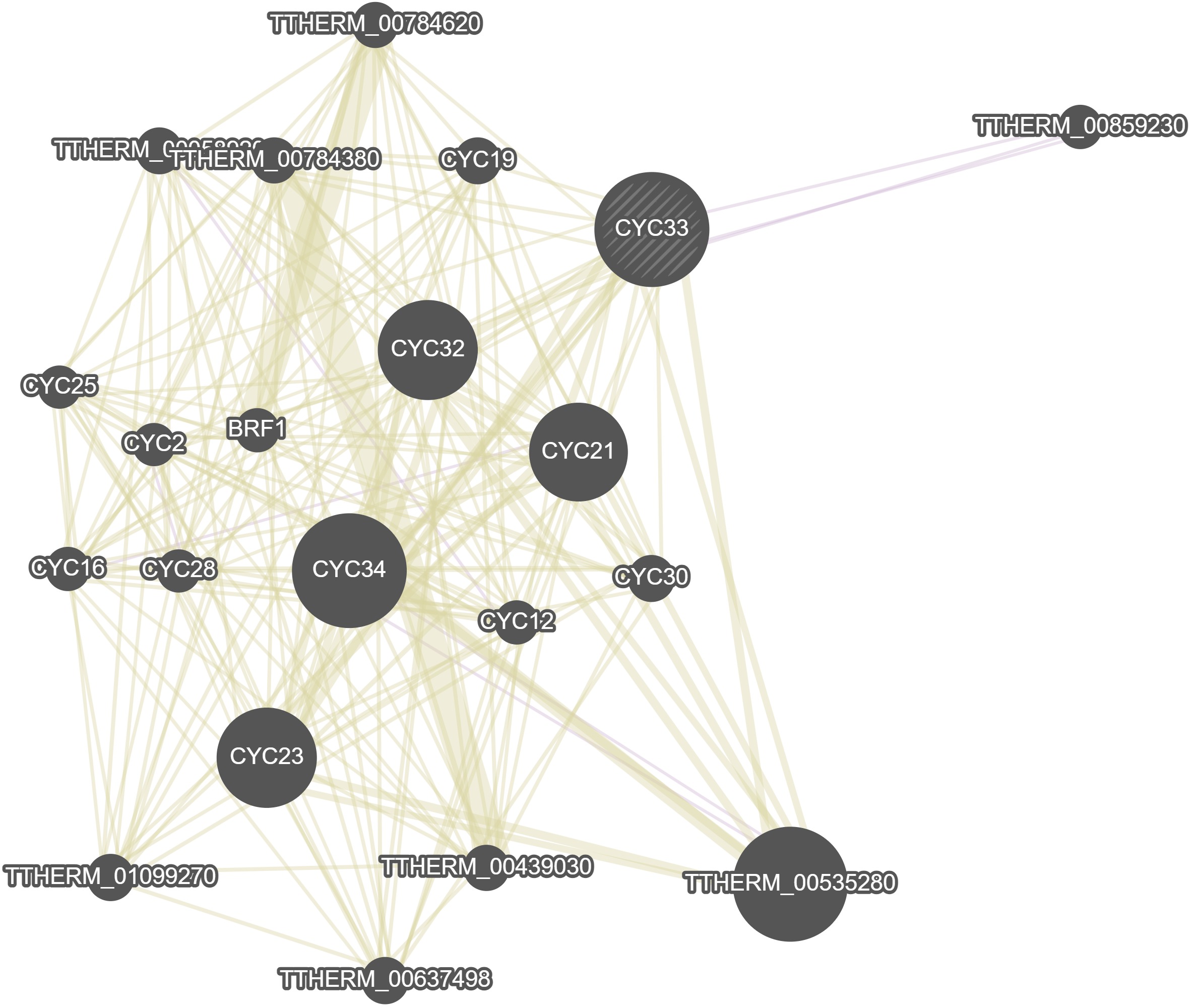
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:27192402: Yan GX, Dang H, Tian M, Zhang J, Shodhan A, Ning YZ, Xiong J, Miao W (2016) Cyc17, a meiosis-specific cyclin, is essential for anaphase initiation and chromosome segregation in Tetrahymena thermophila. Cell cycle (Georgetown, Tex.) 15(14):1855-64
 Sequences
Sequences
>TTHERM_00294860(coding)
ATGACTGATCTATATCATGGAAATTCACCTGAACACTTTTTGAGTACTTGTGGAAGCGAC
AATTTTGATTCTATTGTAGCTAACTCAATCTCAGATAAAAACTGCACCCTTAAAGCCAAG
CAAATGTCATAGAGCTCTTTGTCATCTGTCTCAACTTCATCATCTTCCTTATCACAATCT
TCAATTATTGCTTAAAAGCCTTTAAACGCTGCTAAATATTAGCAAATTATTGAACAATTT
GACAAGCTAGAGAACAGCAATAAATCTAAATCAAGTTCTAAAAGCTCTATTTCATCTTCT
TAAATTTCTTAAAAGAAGATTAATGTTATTAAATTGACTGATTGGGTAGTCAACGACCTT
TGGAAGGATGCCTTTGAGCTCTACTAATTGTAGAATGAAGACGCTGAATACCTTGAGAAT
TGCTCCTTTTATTTAGAAGAAAACAAGCGCATTAAAATCTCATTTGCTTCTTATGTTAAA
AGACTTAAAGAACTAACTGAATGTTCTGATAATTGTTTCATTCTTGCCTTGTTACTCTTC
GATAGACTAAACAAGAAGAAGAAATTAAACTATAGTCGCATCAATGTTCACAAACTTATG
GCCATATGCCTTTGGTTGAGTGTCAAGTTCTATGAAGACATCAATTTCACTGATGCTTAC
TATGCACAAAAGATTGCTGGTATTCCTCTTGAGGAGCTCATTTCTCTTTAATTCGAACTT
CTCGAGCTTTTGAATTATAGATTATTCATTTCACCTCAAAGATTCAACCACTTCATGAAG
CTCATCAACTCTCTCATTCAATCTGGAGCTGTTAATGCTTGA>TTHERM_00294860(gene)
AAGGAAATCTTCCAAAATACAAAATCCTCAAACAAAAAGCAAACTATCCTTGCTAAATTT
TTACCATCTCTTTTATTCATTCAAAAAGAATTTAAAAGTCTCCAAGACTGAGAAAAATCA
TCACAAACAACATAGTTGCGCACTTCTTTTATCTAAATCCAAAGCGCAAGCCTCCTTGAT
AAAACAAACATTCAACTTCATTACTTATCCACTTTAAAATTTAAATCTGATAATTAAAAT
TACAAAAAATGACTGATCTATATCATGGAAATTCACCTGAACACTTTTTGAGTACTTGTG
GAAGCGACAATTTTGATTCTATTGTAGCTAACTCAATCTCAGATAAAAACTGCACCCTTA
AAGCCAAGCAAATGTCATAGAGCTCTTTGTCATCTGTCTCAACTTCATCATCTTCCTTAT
CACAATCTTCAATTATTGCTTAAAAGCCTTTAAACGCTGCTAAATATTAGCAAATTATTG
AACAATTTGACAAGCTAGAGAACAGCAATAAATCTAAATCAAGTTCTAAAAGCTCTATTT
CATCTTCTTAAATTTCTTAAAAGAAGATTAATGTTATTAAATTGACTGATTGGGTAGTCA
ACGACCTTTGGAAGGATGCCTTTGAGCTCTACTAATTGTAGAATGAAGACGCTGAATACC
TTGAGAATTGCTCCTTTTATTTAGAAGAAAACAAGCGCATTAAAATCTCATTTGCTTCTT
ATGTTAAAAGACTTAAAGAACTAACTGAATGTTCTGATAATTGTTTCATTCTTGCCTTGT
TACTCTTCGATAGACTAAACAAGAAGAAGAAATTAAACTATAGTCGCATCAATGTTCACA
AACTTATGGCCATATGCCTTTGGTTGAGTGTCAAGTTCTATGAAGACATCAATTTCACTG
ATGCTTACTATGCACAAAAGATTGCTGGTATTCCTCTTGAGGAGCTCATTTCTCTTTAAT
TCGAACTTCTCGAGCTTTTGAATTATAGATTATTCATTTCACCTCAAAGATTCAACCACT
TCATGAAGCTCATCAACTCTCTCATTCAATCTGGAGCTGTTAATGCTTGAATATCTTCTC
TCAACAACTTTATCCTTTCTATTGTTATTTATGCTCAATTCCAAGAAATTCAAAGCACCT
ATATATAACTTAATATGGAAATTAATAGCAATAATTACAATGTCTTAATCTTAATGGTCT
AAGTAAAGAAATGAATAATTTTAAGAAGAAAAAAGGTATCACATCACATTCAGTCAGTCA
GTATGTTATCTTGCAGATAAATCAACCAAATTTGAGTTAATTTTTTAAGCGTTTCAACGT
ATTGTTTCTTTAATTAGTTTCATCGCTTAGTAATATCATCAGTGTGATTTTATTTATTAA
CTTTTTCAATAATACTGGGTCTTACTTAGAATTGTTAATAAATCACTCTGATCGATTATT
ATGGTTAGCTTGTTTTCATGTTGGTAAATGTTTTTTACGTTTTGAAAACAATTAAAGAAA
AATTAACTCACTTAATCACTCACACACAATTCACTCACTCACTCACTCACTCTTGCCATT
TCAAATTGAAGAACATCATTTTTCTTAAAAATTGTCAAATTTTTGTTAATAAATATATTT
AAACCTAACTAAATTGTTAATAAAACTCTTGTACATAGTTGAAAACTAGCCTTAATATTT
TCACGCTTGAAAAAAGCTAAGCTTCTGTGAATAGTTAGTTCACTTATATGTTGCCTTGCA
ATTTAAATGTTTGAAACTTGTTTATAAATGTTTAAATTAAGCTAGTTAATTGTTATTAAT
AGTCCATTTATCTATAGTATTTTAATATTAAAACAAACTAGTTGATGCAAATTTCATTTA
CATTAACTAGCAGTTTTATCCAATCATGTAAATAAATTATTGAATGATCATCTATAAAAT
TACTAATATTTCAACAAAACCTTATCCCTCTTTGCAATCCCTTCCATCATTCTTTCAATA
AAAATTAAATATTGAAGTTAAGGGCCTCAAAAATGTGCTGTTGAAATCATAGGTCTCAAT
AACAAACGAGTCTTGGAATGAAAGCATGCTAATAACACTAAGAAAAAAGAATAATGAAAA
ATAAATAATTTTAATCAAATACATTATTTAAAAACGAGAATAATTGATAATTTTTTAATA
TAAATTGTTAAATTGAGTTATAATTGAATAATATATAAAATAAATATATAAATTTAATTA
TTTAGTAATTGTATATATTCTTAGAAAATCCATCCATATCTATCTATATTT>TTHERM_00294860(protein)
MTDLYHGNSPEHFLSTCGSDNFDSIVANSISDKNCTLKAKQMSQSSLSSVSTSSSSLSQS
SIIAQKPLNAAKYQQIIEQFDKLENSNKSKSSSKSSISSSQISQKKINVIKLTDWVVNDL
WKDAFELYQLQNEDAEYLENCSFYLEENKRIKISFASYVKRLKELTECSDNCFILALLLF
DRLNKKKKLNYSRINVHKLMAICLWLSVKFYEDINFTDAYYAQKIAGIPLEELISLQFEL
LELLNYRLFISPQRFNHFMKLINSLIQSGAVNA