Gene Model Identifier
TTHERM_00304150
Standard Name
SNF5
Aliases
PreTt21221 | 29.m00182 | 3696.m00005
Description
SNF5 SNF5/SMARCB 1/INI1 family protein; Conserved protein; Member of the SWI/SNF complex; SNF5/SMARCB1/INI1
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
>TTHERM_00304150(coding)
ATGTCTTTAACTGTAACTTATAATAGTGGAATGGTTAAAAAGCCATTATACTAAAGAGTA
CACGAAGTACTTAATCGCTTAGGTTTATTTTATCAAGACATTTGTGTGGAGGCAAATATA
AATGAAGAAAGTTTGATTTAGTGGGTAAACTAAGACCCTGAATATGATTATGCCGAATTA
GAGCTTTCTCTTGAGAAATGGATAGATTCAAAGTACTAGCGTTCATAGAACAATGATGAA
AAATAATATTATTAAATTCGTTCTCAATATTTAAAAAGATTAGATATAAATTAATAGCTA
GCAAACTAATGGCTAGAGGCACCAAATATTGATAAAATTCCTTAAAATGACCAACCAAAT
GAGTATCTAATACCTATAAGCATTGATATTGAATTGGATGGATAGACTTTTAAAGAAAAT
ATAATTTGGAATTATTATGAACCATATTTTACTCCAGAAAACTTTGCTCATCACTTAGCA
AAAGAAAACAGATTAAGTCCTAATATAGAATAAGAAGTAGTTAATGTAATCCGCAGAGCT
CTTTAAAACTATCGCTTCTACGATCCTAAGGAAAGAGAGCTCATCCGTACTATTGAATTA
AATATATTGATAGAAAATGTATAGCTAAAAGATAGATTTGAGTGGGATATTAATGATTTC
ACAAATTCTCCAGAATAATTTGCTTTTAATATGTGCAATGAATTAGGTCTTAGTGGAGAA
TTTGCATAGAGAATAGCGTATTAAATAAGAGAATAGATTCTCTCCTTCTAAAAATAAATT
ATGGAAAGACGTATAAACTAAACTGCATCTAATTTCAAGAAGTCTACTCGTTCTTTAATT
GAATCCTCTTAAAATTTCACAAACTTTGACCCCAAAATTCACATCTAAGGAAATACAGTA
ATTACAGAAGAAAATTATTTACGTCCTGTACTTTAAGATAATGATTATGATAGTTTTCAA
TGGGAGCCAAAAATTTCAATCTTATAACCAGATGATATTAGAAAGGTATAGAAAGAAGAA
TAGAGAAAGCAACGTCTTATGCGTAGAACTCGTTGA
>TTHERM_00304150(gene)
ATAAATTAACAATCTCATTTATTAAAGTATTTAATAAATTAAAAGCTCTTCTATAAGCGG
CAACCATCAAAAATATTATAAAATATAGATTTTAATAAAAGTAGATCATTAGAATATCAA
TAGGATAAAATGTCTTTAACTGTAACTTATAATAGTGGAATGGTTAAAAAGCCATTATAC
TAAAGAGTACACGAAGTACTTAATCGCTTAGGTTTATTTTATCAAGACATTTGTGTGGAG
GCAAATATAAATGAAGAAAGTTTGATTTAGTGGGTAAACTAAGACCCTGAATATGATTAT
GCCGAATTAGAGCTTTCTCTTGAGAAATGGATAGATTCAAAGTACTAGCGTTCATAGAAC
AATGATGAAAAATAATATTATTAAATTCGTTCTCAATATTTAAAAAGATTAGATATAAAT
TAATAGCTAGCAAACTAATGGCTAGAGGCACCAAATATTGATAAAATTCCTTAAAATGAC
CAACCAAATGAGTATCTAATACCTATAAGCATTGATATTGAATTGGATGGATAGACTTTT
AAAGAAAATATAATTTGGAATTATTATGAACCATATTTTACTCCAGAAAACTTTGCTCAT
CACTTAGCAAAAGAAAACAGATTAAGTCCTAATATAGAATAAGAAGTAGTTAATGTAATC
CGCAGAGCTCTTTAAAACTATCGCTTCTACGATCCTAAGGAAAGAGAGCTCATCCGTACT
ATTGAATTAAATATATTGATAGAAAATGTATAGCTAAAAGATAGATTTGAGTGGGATATT
AATGATTTCACAAATTCTCCAGAATAATTTGCTTTTAATATGTGCAATGAATTAGGTCTT
AGTGGAGAATTTGCATAGAGAATAGCGTATTAAATAAGAGAATAGATTCTCTCCTTCTAA
AAATAAATTATGGAAAGACGTATAAACTAAACTGCATCTAATTTCAAGAAGTCTACTCGT
TCTTTAATTGAATCCTCTTAAAATTTCACAAACTTTGACCCCAAAATTCACATCTAAGGA
AATACAGTAATTACAGAAGAAAATTATTTACGTCCTGTACTTTAAGATAATGATTATGAT
AGTTTTCAATGGGAGCCAAAAATTTCAATCTTATAACCAGATGATATTAGAAAGGTATAG
AAAGAAGAATAGAGAAAGCAACGTCTTATGCGTAGAACTCGTTGAGATGTTGAAGCATAT
CATTAAAGATAAGATAAATCATTTATTACATCATAATTTAAAGTGTAATTTTCTAGCACC
AAAACTCTATTTAAGAAGTATTTTAAACATATTGATTGGTAGGTAGGTAGGTATGTCATG
GTTTTATTTATTCATATATAAAAAAAAGTGAATATCATGTATACCACCCTTAAAAAATTG
TATTCCATTTATATTATAAGAATTAGTAATTATTTGAGGAATTTAAAATAAATTTTATGT
TGTCAGTCATAAAAGATTTCGTCAAA
>TTHERM_00304150(protein)
MSLTVTYNSGMVKKPLYQRVHEVLNRLGLFYQDICVEANINEESLIQWVNQDPEYDYAEL
ELSLEKWIDSKYQRSQNNDEKQYYQIRSQYLKRLDINQQLANQWLEAPNIDKIPQNDQPN
EYLIPISIDIELDGQTFKENIIWNYYEPYFTPENFAHHLAKENRLSPNIEQEVVNVIRRA
LQNYRFYDPKERELIRTIELNILIENVQLKDRFEWDINDFTNSPEQFAFNMCNELGLSGE
FAQRIAYQIREQILSFQKQIMERRINQTASNFKKSTRSLIESSQNFTNFDPKIHIQGNTV
ITEENYLRPVLQDNDYDSFQWEPKISILQPDDIRKVQKEEQRKQRLMRRTR
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
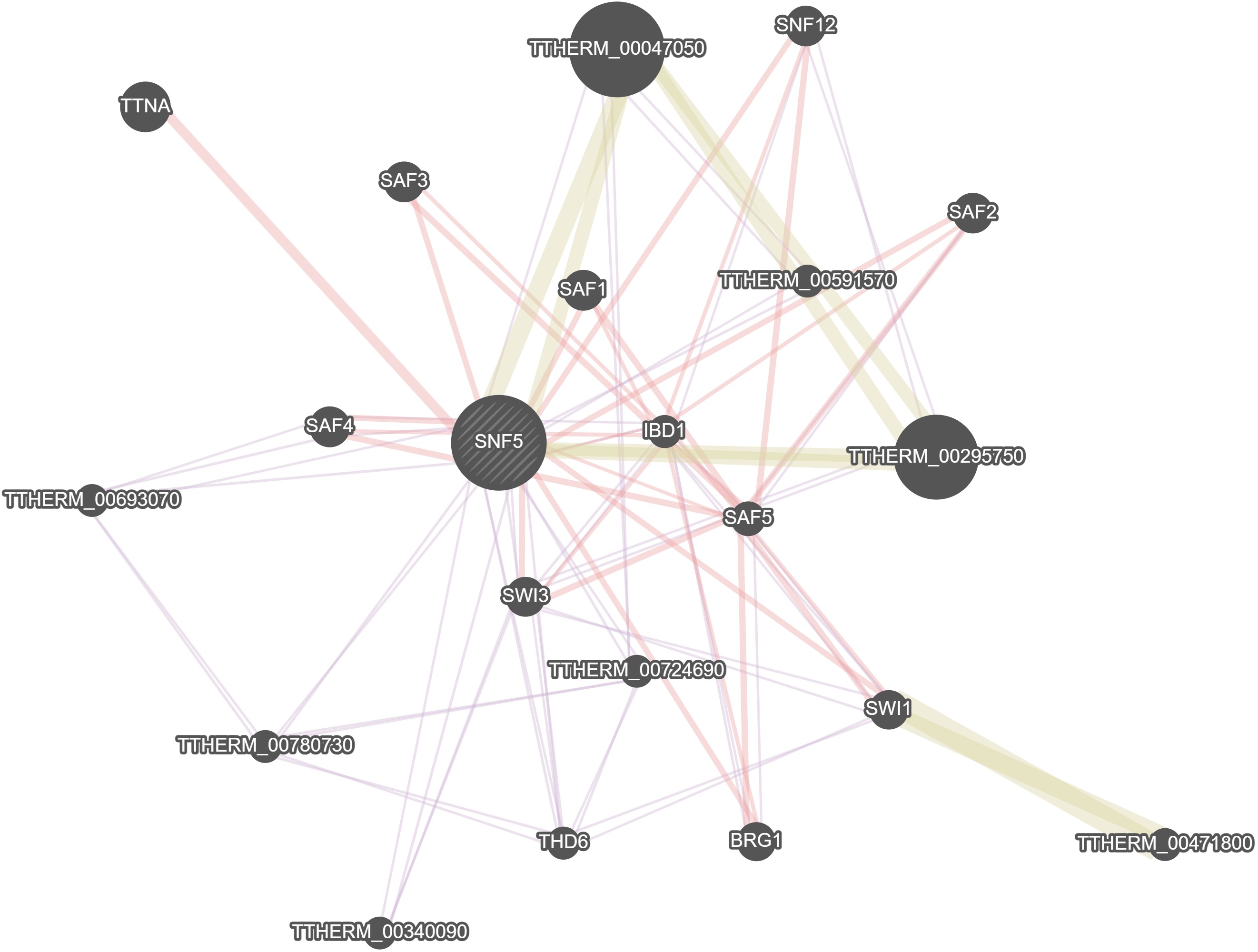
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences