Gene Model Identifier
TTHERM_00585190
Standard Name
HPL5
(Heterochromatin Protein 1 Like 5)
Aliases
PreTt04125 | 80.m00171 | 3675.m00069 | HPC_SH | COI13
Description
HPL5 chromodomain protein; Chromo domain protein; Chromo domain
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
>TTHERM_00585190(coding)
ATGAATAACAAATATTATCATCAACCTTCCTAAAAATTTGAAATCCTGGCCATATTAGAT
AAAAGGGCAAATCCTAAGAATGGGAAAATAGAATATTTAGTCCAGTGGAAATAATGGCCT
CTAGATTATACATGGGAGCCTAAAAAAAACATTTAAAATTTTATTGATCAAAATGAGTTA
TTTGTTGATGATGAAGACCTTAGGAGACATTATAAGGAAAAGCAATATCGAAGAAAAATC
TTAAGAGAAGAAAGAAATAATTAAGTTGAAAAGATATGCAATAGTATAGTTAATCATACT
TAAATTAATACTACTTAGAATAGATAAAATGATAATGATTTACTTAGTAGTATTGATGAT
TATGAAGAAAATATAATATACGAGGAGAGCTTTGATGAATCGAATTATTAAGAAAACTGC
AAGGAATCATGGAGCACAAATCCGAATACATAATAAACTATTAGGAAAGATGAATTTTTT
TTATAAGAACCTGATAGCTATTCTATGAAAAAGATTTAAAATAAAAAAGAGATTAGTTCA
TTAAAAATAGAAAATACTTTTATCAACTCAAAGGCATAAAAGTTTAATAAATCTTCTTCA
GAAATAGATTTAAGTTAAAATGAAGGATCATAAAAAGTTGATTAAATGAATGAGTACTAA
GTTAATTAAGATAATTTTAAAACTGAGGGATTTTCAGAAGAAAATTTATTTGAAAATAAA
TCTGCTTCCACTGAATAAATTCAATTCTCTAATGCTAATGATAGCTAAAAGCATTAAAAA
ATTGTTAGAAAGTATTCAAATGTGATTTCAAAAGATGAAGAAATCGGAGAATTTATGAAA
GATGAAATTTCAAACATAGAAATGGAAATAAAATATTCATTTAAGGTGAATTGGAAAAGA
AGAAGCAATGGAGTCACTCCATTGTCAAAAACAATAGAAGTTCTTAGTAAAAAGCATATT
TCTCCTTAAGAAATGATTGATTATTTAACTTAATCAAATTAAATTAATAATGAAATAGAT
AAAAGTTAAAAATGA
>TTHERM_00585190(gene)
CTAATTTTATGTATAATTTACAAAAACCATTCTAATAATTTTATTTTTCAATATTTATTA
TCATTTTATCTATTTTTGAAATTATACAAAAGAATCATTAATAATCTAATATAATCTAAA
ATTATTAATAAAGAATTTCTAAATATATTAAGTTCTACTAAAAAATAAAATGAATAACAA
ATATTATCATCAACCTTCCTAAAAATTTGAAATCCTGGCCATATTAGATAAAAGGGCAAA
TCCTAAGAATGGGAAAATAGAATATTTAGTCCAGTGGAAATAATGGCCTCTAGATTATAC
ATGGGAGCCTAAAAAAAACATTTAAAATTTTATTGATCAAAATGAGTTATTTGTTGATGA
TGAAGACCTTAGGAGACATTATAAGGAAAAGCAATATCGAAGAAAAATCTTAAGAGAAGA
AAGAAATAATTAAGTTGAAAAGATATGCAATAGTATAGTTAATCATACTTAAATTAATAC
TACTTAGAATAGATAAAATGATAATGATTTACTTAGTAGTATTGATGATTATGAAGAAAA
TATAATATACGAGGAGAGCTTTGATGAATCGAATTATTAAGAAAACTGCAAGGAATCATG
GAGCACAAATCCGAATACATAATAAACTATTAGGAAAGATGAATTTTTTTTATAAGAACC
TGATAGCTATTCTATGAAAAAGATTTAAAATAAAAAAGAGATTAGTTCATTAAAAATAGA
AAATACTTTTATCAACTCAAAGGCATAAAAGTTTAATAAATCTTCTTCAGAAATAGATTT
AAGTTAAAATGAAGGATCATAAAAAGTTGATTAAATGAATGAGTACTAAGTTAATTAAGA
TAATTTTAAAACTGAGGGATTTTCAGAAGAAAATTTATTTGAAAATAAATCTGCTTCCAC
TGAATAAATTCAATTCTCTAATGCTAATGATAGCTAAAAGCATTAAAAAATTGTTAGAAA
GTATTCAAATGTGATTTCAAAAGATGAAGAAATCGGAGAATTTATGAAAGATGAAATTTC
AAACATAGAAATGGAAATAAAATATTCATTTAAGGTGAATTGGAAAAGAAGAAGCAATGG
AGTCACTCCATTGTCAAAAACAATAGAAGTTCTTAGTAAAAAGCATATTTCTCCTTAAGA
AATGATTGATTATTTAACTTAATCAAATTAAATTAATAATGAAATAGATAAAAGTTAAAA
ATGAATCAATGATTAATAAAATTATTTTTATTTCTCTGCATAAAAATATAATTAAACTAA
TTAAGTAGACACTTTTCATAGTATCATAATTTTATCTTTAAATTTAGAAAAAGTGACTGG
CTGATCGATTATTTATAAATATTTGTTTTATTTGTTCTCATTTGTAATGTAAATTTGACT
CATTAAAATACCTAATAATAAATTTAATATCTTTTTATTTCTCGATCAATATTTAGATTG
AGACACAAAAATTTTAGAAAAAGTAAAATTTGCATCTACAGTTAATTAATTA
>TTHERM_00585190(protein)
MNNKYYHQPSQKFEILAILDKRANPKNGKIEYLVQWKQWPLDYTWEPKKNIQNFIDQNEL
FVDDEDLRRHYKEKQYRRKILREERNNQVEKICNSIVNHTQINTTQNRQNDNDLLSSIDD
YEENIIYEESFDESNYQENCKESWSTNPNTQQTIRKDEFFLQEPDSYSMKKIQNKKEISS
LKIENTFINSKAQKFNKSSSEIDLSQNEGSQKVDQMNEYQVNQDNFKTEGFSEENLFENK
SASTEQIQFSNANDSQKHQKIVRKYSNVISKDEEIGEFMKDEISNIEMEIKYSFKVNWKR
RSNGVTPLSKTIEVLSKKHISPQEMIDYLTQSNQINNEIDKSQK
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
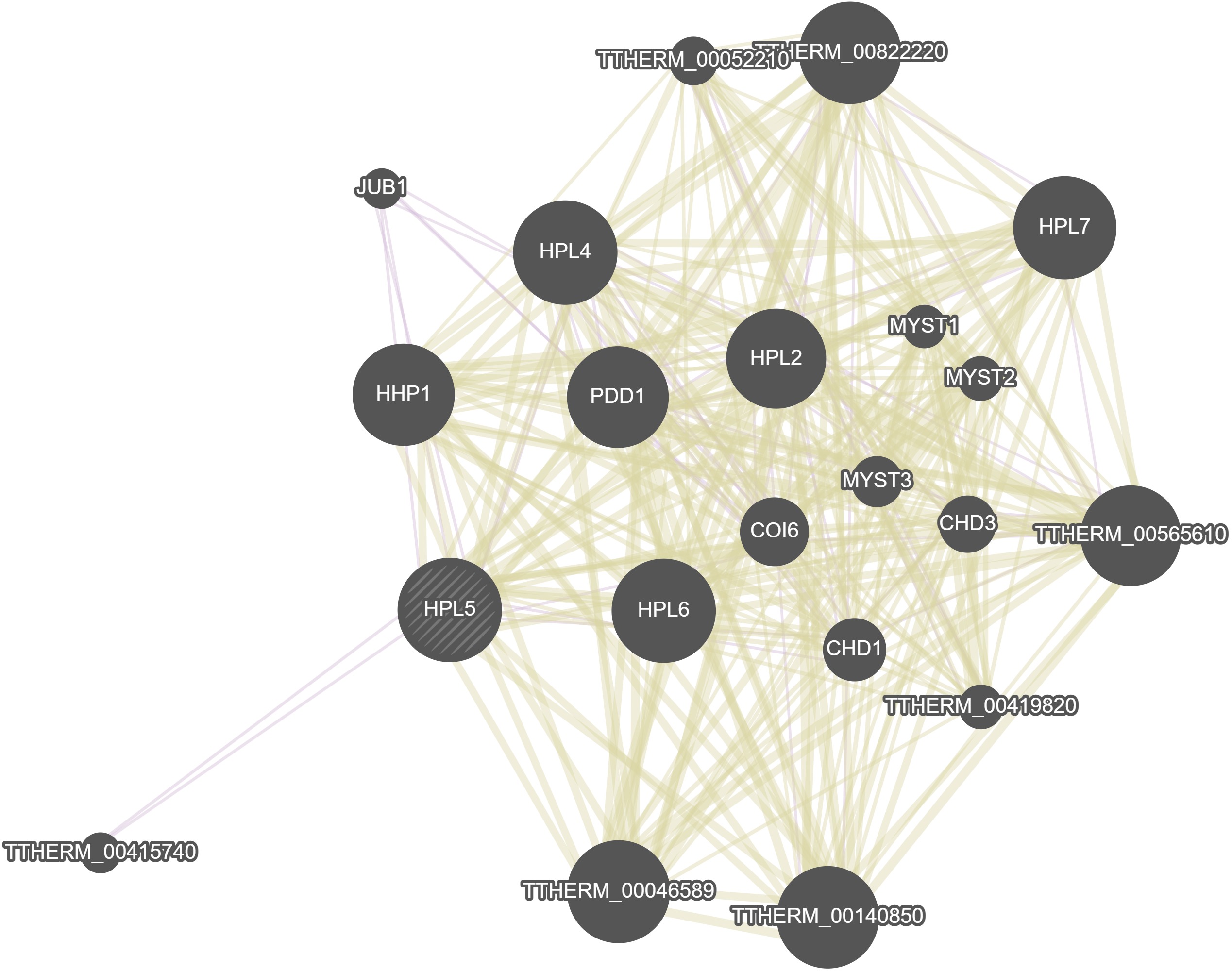
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences