Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00664060Standard Name
RAB41 (RAB GTPase 41)Aliases
PreTt23853 | 98.m00196 | 3736.m00094Description
RAB41 small guanosine triphosphatase family (GTPase)-like Ras family protein; Novel Rab family protein; small monomeric GTPase that regulates membrane fusion and fission eventsGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- intracellular anatomical structure (IEA) | GO:0005622
Molecular Function
- GTP binding (IEA) | GO:0005525
Biological Process
- small GTPase-mediated signal transduction (IEA) | GO:0007264
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
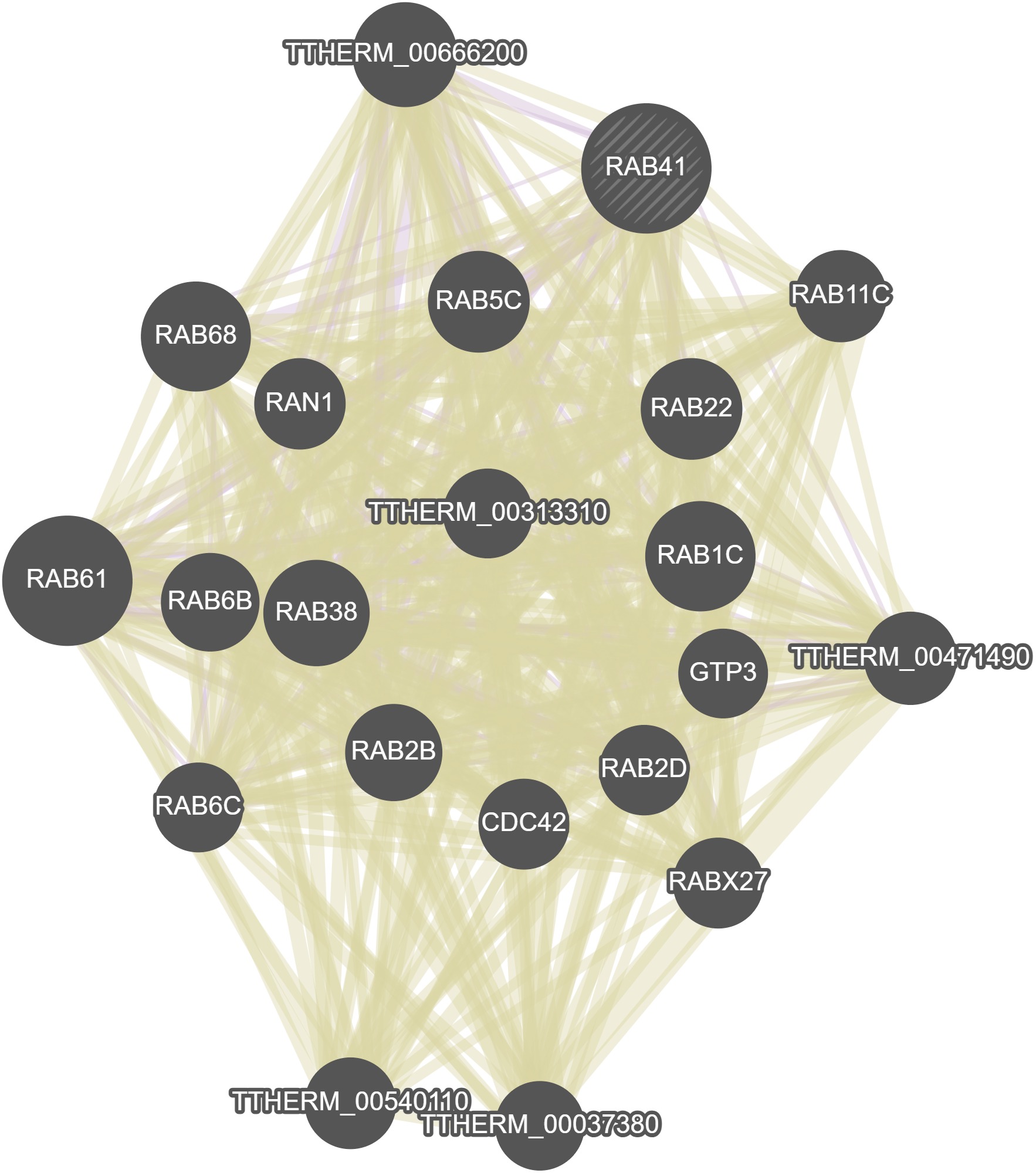
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
Source Identifier Score Tetrahymena borealis EI9_17189.1 2.0000211193600847e-137 Description: Ras family protein (238 aa) Oxytricha Contig15751.0.0.g87 1.0001801308941382e-55 Description: Ras family DictyBase DDB_G0271736 9.998397555368434e-52 Description: rabC on chromosome: 2 position 718547 to 719569 WormBase WBGene00004266 8.003782782428492e-46 Description: locus:rab-1 status:Confirmed UniProt:Q9UAQ6 protein_id:CCD67094.1 SGD YFL038C 1.000159011384939e-43 Description: YPT1 Rab family GTPase, involved in the ER-to-Golgi step of the secretory pathway; complex formation with the Rab escort protein Mrs6p is required for prenylation of Ypt1p by protein geranylgeranyltransferase type II (Bet2p-Bet4p) Stentor Coeruleus SteCoe_14213 1.9287498479639178e-22 Description: None
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:16933976: Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA,
 Sequences
Sequences
>TTHERM_00664060(coding)
ATGTAAAAAGAAAATAGTGGGGTTTATTCAAATTAGCTTTAAGCTGGAGTTTTTAATTCC
CCACACACGAAGATTGACTTAGTTTTTAAAATAATTTTAATAGGACAGCAAAATGTAGGC
AAATCTTCCATTTTAAATTAGTACGTAGATAGAGAATTTGCAGAACATTATATGTCAACA
ATAGGTGTAGATTTTAGATTAAAAACAATTAAAACAAGTTCTGGTAAAATAGTGAAGCTA
CAAATTTGGGATACTGCTGGATAAGAAAGATTTGCAGCTTTGACTAATTCACATTATAAA
GGAGCGCATTCTTGCATTTGTGTCTATGATATAACTAATCCTAAGAGCTTAGCATCAGCA
GAATAATATTTATAGACAACAATTAACCAACATGGGATAGATTCAGATTTAATTTATCTG
ATAGGCAACAAATCAGATTTAGAAGATTAAAGGAAGATTAGCGAAAAAGAAGGAAAAGAT
TTAAGTTATAAATACGGAGTAAATTTCTTAGAAACAAGTGCAAAAAATTACTTAAATATA
GAAGAGCTTTTTGATTAGATGGTTGAGCACTTAGTGAGTCTAATTACTAACCCTCAAATA
CCAAAAAAACTTTCATAGTTAAATAATGTCAAAATAACTAGCAAAGATATTATGATTAAA
TAGTAAGAAGGTAATAATGAAGTTTAAAAGAAAGGCTGCTGTTGA>TTHERM_00664060(gene)
ATGTAAAAAGAAAATAGTGGGGTTTATTCAAATTAGCTTTAAGCTGGAGTTTTTAATTCC
CCACACACGAAGATTGACTTAGTTTTTAAAATAATTTTAATAGGACAGCAAAATGTAGGC
AAATCTTCCATTTTAAATTAGTACGTAGATAGAGAATTTGCAGAACATTATATGTCAACA
ATAGGTGTAGATTTTAGATTAAAAACAATTAAAACAAGTTCTGGTAAAATAGTGAAGCTA
CAAATTTGGGATACTGCTGGATAAGAAAGATTTGCAGCTTTGACTAATTCACATTATAAA
GGAGCGCATTCTTGCATTTGTGTCTATGATATAACTAATCCTAAGAGCTTAGCATCAGCA
GAATAATATTTATAGACAACAATTAACCAACATGGGATAGATTCAGATTTAATTTATCTG
ATAGGCAACAAATCAGATTTAGAAGATTAAAGGAAGATTAGCGAAAAAGAAGGAAAAGAT
TTAAGTTATAAATACGGAGTAAATTTCTTAGAAACAAGTGCAAAAAATTACTTAAATATA
GAAGAGCTTTTTGATTAGATGGTTGAGCACTTAGTGAGTCTAATTACTAACCCTCAAATA
CCAAAAAAACTTTCATAGTTAAATAATGTCAAAATAACTAGCAAAGATATTATGATTAAA
TAGTAAGAAGGTAATAATGAAGTTTAAAAGAAAGGCTGCTGTTGA>TTHERM_00664060(protein)
MQKENSGVYSNQLQAGVFNSPHTKIDLVFKIILIGQQNVGKSSILNQYVDREFAEHYMST
IGVDFRLKTIKTSSGKIVKLQIWDTAGQERFAALTNSHYKGAHSCICVYDITNPKSLASA
EQYLQTTINQHGIDSDLIYLIGNKSDLEDQRKISEKEGKDLSYKYGVNFLETSAKNYLNI
EELFDQMVEHLVSLITNPQIPKKLSQLNNVKITSKDIMIKQQEGNNEVQKKGCC