Gene Model Identifier
TTHERM_00786930
Standard Name
NAP1
(Nucleosome Assembly Protein )
Aliases
PreTt08520 | 132.m00079 | 3755.m00440 | g2363
Description
NAP1 nucleosome assembly protein; nucleosome assembly protein (NAP); Nucleosome assembly protein (NAP)
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
>TTHERM_00786930(coding)
ATGTCTTCTGAAACCGCCGCCAAGCACGACCACTAAAACTGTGAAGATCACGATCACGAT
CACGAAGAAATCGACATCGATTAATAAATCGAAGAATCCATCAAGGAATTAGCTCTTGGT
GAAAAGATCAGAGCTGTTGCTATCAACCACCACCTCGCTGAAAAGAAGGCCCTCGATGAA
GAACTCTAAAAGAAGATCCAAGCCCTCACCTTCGAATACGAACAAAAATCCATGCCCATC
TACGCTAAGACTCAAGAAATCGTTCAAGGTAGAGTTCCCACTGAAGAAGAATTGAAGGAC
TTAGAAAAGTACTTAAAGGAAGAAGAAAAGACTCAAGTTGAAGGTGCCAAGAAGGAAGAA
CCCATCACCGAATATTGGTTGAAGGCTATGAAGAGCAACGACGTTTTGGCTATGGAAATC
AAGGAACAAGACGAAACTGCCTTAAAGACCCTTACCAAGATCGAATACGTTTTGGAAGAT
ACCAAGAAGTTCCACATCGTCTTCACTTTCGGTCCCAACGACTACTTCACCAACACTGAA
TTAAAGAAGACCGTCGAATTAGATGAAAACGAAGAACCTGTTAAGACCACCGGTACTCCC
ATTGAATGGAAGGAAGGAAAGAACACCACCGTCAAGATCACCAAGAAGACCCAAAAGAAC
AAAAAGACTGGTGTCAAGAGAGTCGTTGAAAGAGAAACCAAGATCGAATCTTTCTTCAAT
TTCTTCTCTGACTCTGCTCCCACTGATGCCCCCGAAGAAGAAGACGAAGAAAAGGACGAA
CAAGCCATGGACGGTGACAGAATCAACATCGACTTCGATATTGCTAGATCTTTAATCGAT
GAAGTTATTCCCTACTCTCTTGAATACTTCCTTGGTATTAAGACTGGTGGTGAAGATCAC
GATGAAGAAATCGATATGGATGACCTCGATGAAGAAGAAATCGCCTAATTGCAAAAGGAA
TACGCTGCCAAGACTGGCAAGAAGGGTCCTGCTGCTGGTGGTGCTAACGATAAGAAGGAC
TGCAAGCAATAGTGA
>TTHERM_00786930(gene)
AAAAAAATTAAAATCTGATTAAAATTAATAAAGCAAATAATCTTAAAAGCAATAAAAACA
AATCTTTCTAAAATGTCTTCTGAAACCGCCGCCAAGCACGACCACTAAAACTGTGAAGAT
CACGATCACGATCACGAAGAAATCGACATCGATTAATAAATCGAAGAATCCATCAAGGAA
TTAGCTCTTGGTGAAAAGATCAGAGCTGTTGCTATCAACCACCACCTCGCTGAAAAGAAG
GCCCTCGATGAAGAACTCTAAAAGAAGATCCAAGCCCTCACCTTCGAATACGAACAAAAA
TCCATGCCCATCTACGCTAAGACTCAAGAAATCGTTCAAGGTAGAGTTCCCACTGAAGAA
GAATTGAAGGACTTAGAAAAGTACTTAAAGGAAGAAGAAAAGACTCAAGTTGAAGGTGCC
AAGAAGGAAGAACCCATCACCGAATATTGGTTGAAGGCTATGAAGAGCAACGACGTTTTG
GCTATGGAAATCAAGGAACAAGACGAAACTGCCTTAAAGACCCTTACCAAGATCGAATAC
GTTTTGGAAGATACCAAGAAGTTCCACATCGTCTTCACTTTCGGTCCCAACGACTACTTC
ACCAACACTGAATTAAAGAAGACCGTCGAATTAGATGAAAACGAAGAACCTGTTAAGACC
ACCGGTACTCCCATTGAATGGAAGGAAGGAAAGAACACCACCGTCAAGATCACCAAGAAG
ACCCAAAAGAACAAAAAGACTGGTGTCAAGAGAGTCGTTGAAAGAGAAACCAAGATCGAA
TCTTTCTTCAATTTCTTCTCTGACTCTGCTCCCACTGATGCCCCCGAAGAAGAAGACGAA
GAAAAGGACGAACAAGCCATGGACGGTGACAGAATCAACATCGACTTCGATATTGCTAGA
TCTTTAATCGATGAAGTTATTCCCTACTCTCTTGAATACTTCCTTGGTATTAAGACTGGT
GGTGAAGATCACGATGAAGAAATCGATATGGATGACCTCGATGAAGAAGAAATCGCCTAA
TTGCAAAAGGAATACGCTGCCAAGACTGGCAAGAAGGGTCCTGCTGCTGGTGGTGCTAAC
GATAAGAAGGACTGCAAGCAATAGTGATTGATTTAGTTAATATAAAAAATTATATAGTAA
AGCTATCTCTTGTTTTCTTTCTCACTCTCAATAAACAAACATACATCTGTTATATAGCAA
GCAAACAAAATCATATAGTGAAAGATAAATCTTAAATTATAATTATTATGGAATGAAAGT
AATAATAAAGCATTGATACTAACTTTGCATAATAACTAACAAACAAACAATAACAATAAT
AATTCTTTCGTGTTTTATCTCTATTAGCAATTAATTCAAGTATTAATATAGATAAAAATA
TAACCATGCAAAAAATAATATATCTTTATCAATACCTCTAACTAGAGGACAAGTAAAACG
TACTTCTGTTTATATGCAAATTTGTATTAAA
>TTHERM_00786930(protein)
MSSETAAKHDHQNCEDHDHDHEEIDIDQQIEESIKELALGEKIRAVAINHHLAEKKALDE
ELQKKIQALTFEYEQKSMPIYAKTQEIVQGRVPTEEELKDLEKYLKEEEKTQVEGAKKEE
PITEYWLKAMKSNDVLAMEIKEQDETALKTLTKIEYVLEDTKKFHIVFTFGPNDYFTNTE
LKKTVELDENEEPVKTTGTPIEWKEGKNTTVKITKKTQKNKKTGVKRVVERETKIESFFN
FFSDSAPTDAPEEEDEEKDEQAMDGDRINIDFDIARSLIDEVIPYSLEYFLGIKTGGEDH
DEEIDMDDLDEEEIAQLQKEYAAKTGKKGPAAGGANDKKDCKQQ
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
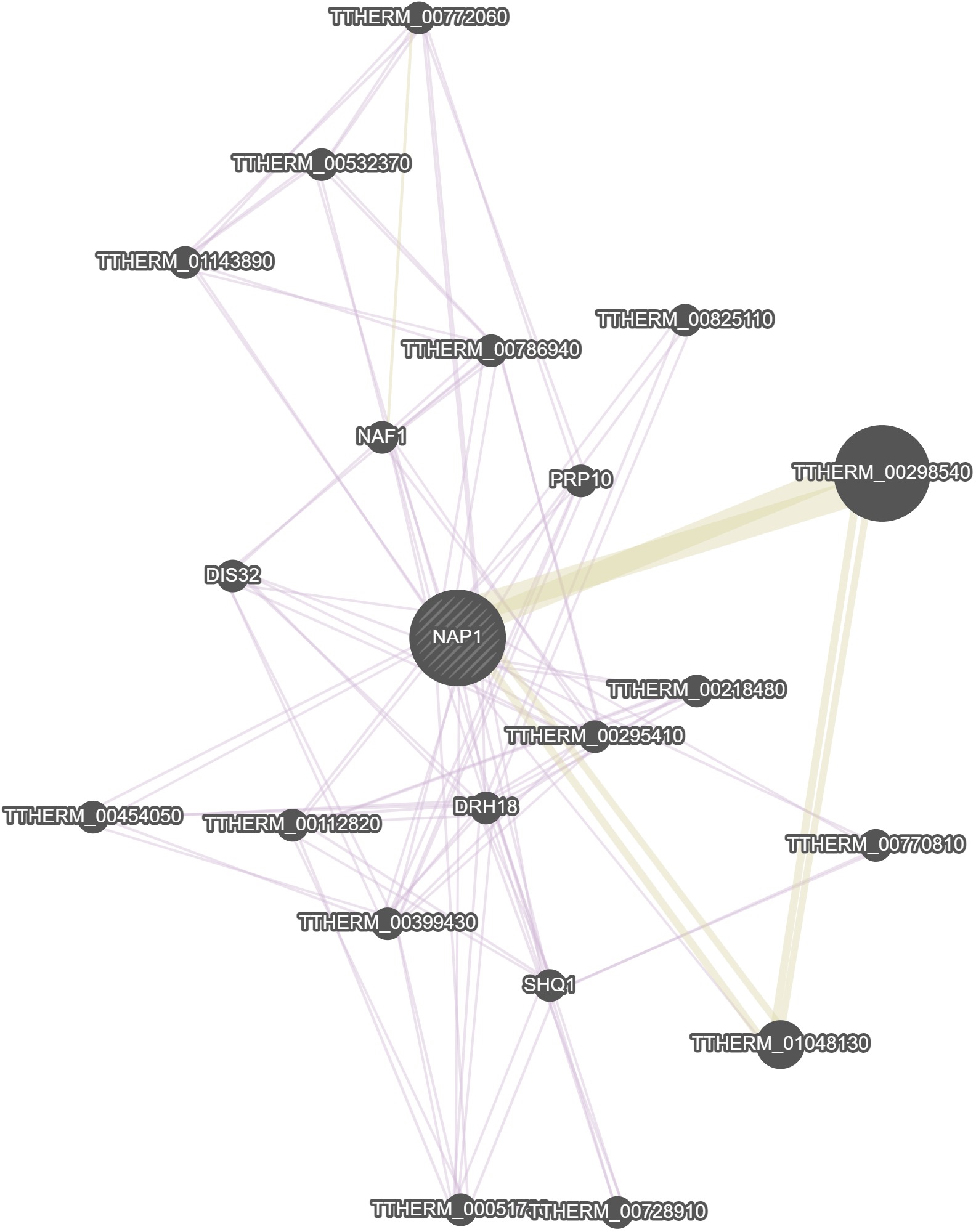
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences