Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_01002860Standard Name
CCF3 (C-terminal Crystallin Fold 3)Aliases
PreTt17206 | 191.m00066 | 3761.m00258Description
CCF3 carboxy-terminal crystallin fold protein; C-terminal crystallin fold protein; domain structure and protein isolation suggest localization in dense core granules; contains additional tandem copies of a 150-residue motif shared by CCF3- CCF4- GRT1- and GRT2Genome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- dense core granule (IDA) | GO:0031045
 Domains
Domains
No Data fetched for Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
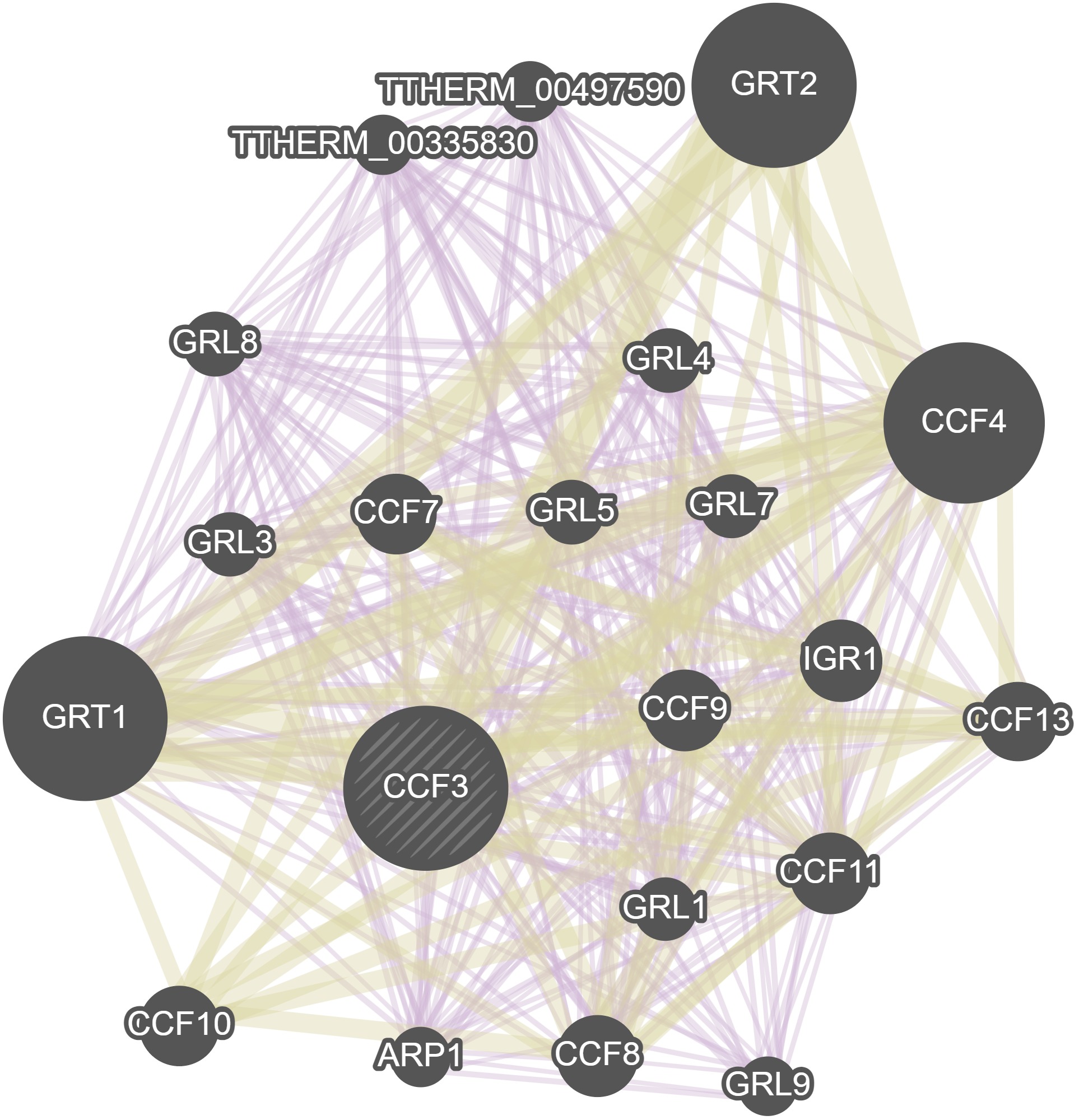
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
Source Identifier Score Tetrahymena borealis EI9_19854.1 9.997093137033517e-262 Description: hypothetical protein (437 aa)
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:16014006: Bowman GR, Smith DG, Michael Siu KW, Pearlman RE, Turkewitz AP (2005) Genomic and proteomic evidence for a second family of dense core granule cargo proteins in Tetrahymena thermophila. The Journal of eukaryotic microbiology 52(4):291-7
 Sequences
Sequences
>TTHERM_01002860(coding)
ATGTAAAAAGGATACATCTTAGTAGCCTTACTTTTAGTAGGTTTTGCTTCCGCTAACTTC
TACGAAGTTGCCAAGGAATTCACTCTCAATAACTTCAATTCCAAATCTCTTAGCAGTTGG
AATCTTGTTAACTTCAAGGGAGATTAAGTCGCCAATTGCAATGGTGTTTCTGTTGTTGGT
GGTCCCAGCGTCGTTGCCAGTGGTTCAATGACCAAAACTTTCACAACTGATTTTGATCTC
AACGGAGTTAATATCTCTTTCGACTTATATTTCTTTGATAAATGGTTCTCTGGTTGGGAT
GGTGGTAGAGATTAGAACACCTTGTTCAAGATCTTCGTTAATAACCAATAAGTTTACCAA
CTTGGTTCTGATTACACCAGATCCCTCCATGCTGATTAACCCAATCTTTGTGGTGATGCT
ACTCCTGATAGATAAACTCACATTTCTCTTGATGTCCCCGCTACTGGTAGAAACATCACT
GTCTAACTTGTTGCCAACATCGCTTCCAATAATCCTGCAGTTGCCTCTTGGGGTATCAGA
AATTTCTACATTTTATCATAATAAGGTTATGATGATTCTTAAGTTGTTGTTAGTGAACTC
AATGATGATTCCAATTTCGATGTTTCTCAATGGTAAACAAATTTCAAAGGTGTAATCACA
TAATGCCCTTCAAAGCTCCTTGGTGGTTACAATGCTGCTGGACCTAAAGCCACTGCTGTT
AGAAAACTCGCAAGTCTCCCTGCTCACTCCTTCTTGACTGTTTCCCTCAGTGCTTATTTC
ATTGATTCCATTGATGCTAATGATTCAGTTGTTATTTACGTTGATGGAAAGGTTGCCCAT
GAACACTAAAAGGCTTATTATGAAGATAGTACTAATGTTTGCGGTGCTGCCTGGAACGAC
GGTGTTGATAATGAATTAAATATCATTGTTCCTCACTTTGCTTCTACTGCTGAAATCAAA
ATTTACGTCAATACTGACCAAGCTCCTGGTGATGAATCCTTCGGTTTCAGAGAATTCTAA
GTTTCTGTTTCTTCTCCCTGTCCTATTCTCTACACTGATGTTAATCAAGGAGGTAGACAA
TTCTCCTTCTGTGGTAATCTTCCTGATCTCAAGGCTGTTGGTTGGACCGCTCCTATTAAA
GGTATTTACCTTCCCAAGGGAGTTGAACTCACTGTCTACTAAGATTCTAACTACAATGGA
AAGAAATACACTGTTAAGAAATCTAACTTCAATCTCTCAGGTAATGAATACTAATTGATT
AACAAAGCTTTAAACATTGATGTTACTGAATAAAAGCTTAGACGTAACAAGTGA>TTHERM_01002860(gene)
ATGTAAAAAGGATACATCTTAGTAGCCTTACTTTTAGTAGGTTTTGCTTCCGCTAACTTC
TACGAAGTTGCCAAGGAATTCACTCTCAATAACTTCAATTCCAAATCTCTTAGCAGTTGG
AATCTTGTTAACTTCAAGGGAGATTAAGTCGCCAATTGCAATGGTGTTTCTGTTGTTGGT
GGTCCCAGCGTCGTTGCCAGTGGTTCAATGACCAAAACTTTCACAACTGATTTTGATCTC
AACGGAGTTAATATCTCTTTCGACTTATATTTCTTTGATAAATGGTTCTCTGGTTGGGAT
GGTGGTAGAGATTAGAACACCTTGTTCAAGATCTTCGTTAATAACCAATAAGTTTACCAA
CTTGGTTCTGATTACACCAGATCCCTCCATGCTGATTAACCCAATCTTTGTGGTGATGCT
ACTCCTGATAGATAAACTCACATTTCTCTTGATGTCCCCGCTACTGGTAGAAACATCACT
GTCTAACTTGTTGCCAACATCGCTTCCAATAATCCTGCAGTTGCCTCTTGGGGTATCAGA
AATTTCTACATTTTATCATAATAAGGTTATGATGATTCTTAAGTTGTTGTTAGTGAACTC
AATGATGATTCCAATTTCGATGTTTCTCAATGGTAAACAAATTTCAAAGGTGTAATCACA
TAATGCCCTTCAAAGCTCCTTGGTGGTTACAATGCTGCTGGACCTAAAGCCACTGCTGTT
AGAAAACTCGCAAGTCTCCCTGCTCACTCCTTCTTGACTGTTTCCCTCAGTGCTTATTTC
ATTGATTCCATTGATGCTAATGATTCAGTTGTTATTTACGTTGATGGAAAGGTTGCCCAT
GAACACTAAAAGGCTTATTATGAAGATAGTACTAATGTTTGCGGTGCTGCCTGGAACGAC
GGTGTTGATAATGAATTAAATATCATTGTTCCTCACTTTGCTTCTACTGCTGAAATCAAA
ATTTACGTCAATACTGACCAAGCTCCTGGTGATGAATCCTTCGGTTTCAGAGAATTCTAA
GTTTCTGTTTCTTCTCCCTGTCCTATTCTCTACACTGATGTTAATCAAGGAGGTAGACAA
TTCTCCTTCTGTGGTAATCTTCCTGATCTCAAGGCTGTTGGTTGGACCGCTCCTATTAAA
GGTATTTACCTTCCCAAGGGAGTTGAACTCACTGTCTACTAAGATTCTAACTACAATGGA
AAGAAATACACTGTTAAGAAATCTAACTTCAATCTCTCAGGTAATGAATACTAATTGATT
AACAAAGCTTTAAACATTGATGTTACTGAATAAAAGCTTAGACGTAACAAGTGA>TTHERM_01002860(protein)
MQKGYILVALLLVGFASANFYEVAKEFTLNNFNSKSLSSWNLVNFKGDQVANCNGVSVVG
GPSVVASGSMTKTFTTDFDLNGVNISFDLYFFDKWFSGWDGGRDQNTLFKIFVNNQQVYQ
LGSDYTRSLHADQPNLCGDATPDRQTHISLDVPATGRNITVQLVANIASNNPAVASWGIR
NFYILSQQGYDDSQVVVSELNDDSNFDVSQWQTNFKGVITQCPSKLLGGYNAAGPKATAV
RKLASLPAHSFLTVSLSAYFIDSIDANDSVVIYVDGKVAHEHQKAYYEDSTNVCGAAWND
GVDNELNIIVPHFASTAEIKIYVNTDQAPGDESFGFREFQVSVSSPCPILYTDVNQGGRQ
FSFCGNLPDLKAVGWTAPIKGIYLPKGVELTVYQDSNYNGKKYTVKKSNFNLSGNEYQLI
NKALNIDVTEQKLRRNK