Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00193660Standard Name
VMA4 (Vacuolar Membrane Atpase)Aliases
PreTt06649 | 16.m00305Description
VMA4 vacuolar ATP synthase subunit E; vacuolar ATP synthaseGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- proton-transporting two-sector ATPase complex, catalytic domain (IEA) | GO:0033178
Molecular Function
- proton-transporting ATPase activity, rotational mechanism (IEA) | GO:0046961
Biological Process
- proton motive force-driven ATP synthesis (IEA) | GO:0015986
 Domains
Domains
- ( PF01991 ) ATP synthase (E/31 kDa) subunit
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
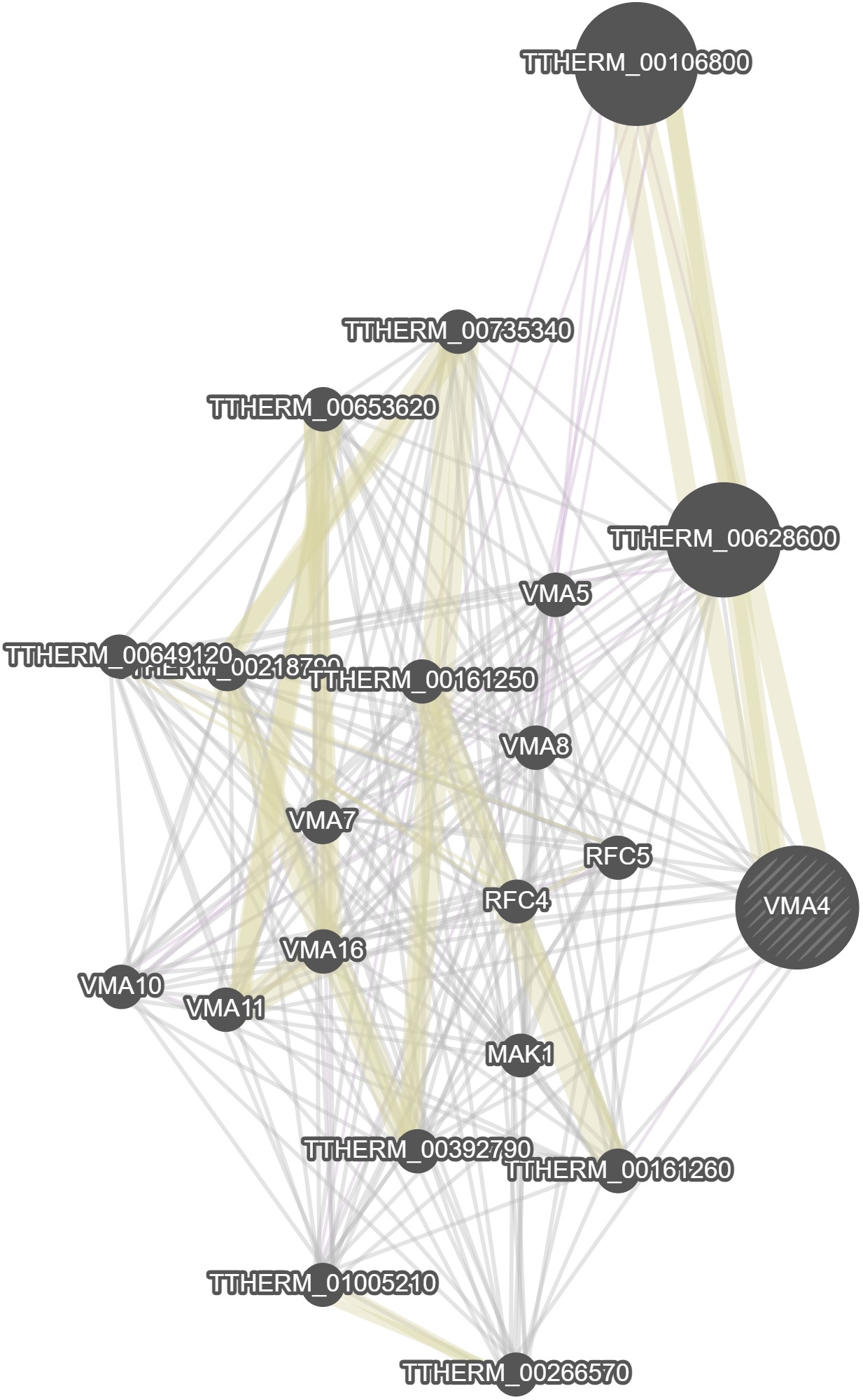
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
Source Identifier Score Tetrahymena borealis EI9_03646.1 6.001368665888701e-135 Description: vacuolar ATP synthase (230 aa) Oxytricha Contig16360.0.g4 2.999884853455273e-42 Description: ATP synthase (E/31 kDa) subunit WormBase WBGene00006917 6.001249495680822e-29 Description: locus:vha-8 status:Confirmed UniProt:Q95X44 protein_id:CCD65002.1 SGD YOR332W 1.9993007828132465e-27 Description: VMA4 Subunit E of the eight-subunit V1 peripheral membrane domain of the vacuolar H+-ATPase (V-ATPase), an electrogenic proton pump found throughout the endomembrane system; required for the V1 domain to assemble onto the vacuolar membrane DictyBase DDB_G0275701 3.001290106883431e-24 Description: vatE on chromosome: 2 position 5679309 to 5680331 Stentor Coeruleus SteCoe_21951 0.0000000000006914400106940203 Description: None
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:28087716: Cheng CY, Young JM, Lin CG, Chao JL, Malik HS, Yao MC (2016) The piggyBac transposon-derived genes TPB1 and TPB6 mediate essential transposon-like excision during the developmental rearrangement of key genes in Tetrahymena thermophila. Genes & development 30(24):2724-2736
 Sequences
Sequences
>TTHERM_00193660(coding)
ATGGCAGATTTTATTAATAAACCTCAAGAAAACGTTTAGAAGATCGTTGAAGCCATCAAA
TCTGAAGCAGAAGAGAAGGCTGAACAAATTAAAAAGAATGCTGAAGAATAATTTAAGATT
CAAAAAAATAATATTGTCAATACTGAAAAGGATAAAATCATTGAAGAATATAAAAAGAGA
TTAGAAAAGCTCATAGTTGACAGAAGAATTCAACGTTCTGCCAAAATCAATGAATAGAGA
TTAGAAAAAATGAAAGCTCGTTTTGACTTTATTGAAAAGCTAAAGGGTGAAATTTCTAAT
AAAATCGTTTAATCCGTTTCAGACCCTAATAAATATAAAAATGTCTTCAAGCAACTTATA
ATCTAGGCCTTGATCAAGTTGATGGAACCTAAGGTTGAGTTAAAAGTAATGAAGAAAGAT
TTGTAGCTTGCTCGTGAAGTAAAGACAGAATGTGAAAATGAATTCAAGGCAATTGCTAAG
AGAGAATGTAATAGAGACTTTAACTGTACTATTATTATAAATGAATACCACTCTCTTGAA
GAAGAAAATCCTAAAGTTATTGGAGGTATTGTTCTCACTTGCGATGGTGGTAGAATTTAG
GTAAACAACACTTTAAATGCAAGAGTTGATCTTGCTTTCTAAGAATTCCTTCCTGACATC
AGAAGAATTCTTTTCCCTTCTATGAAATGA>TTHERM_00193660(gene)
ATGGCAGATTTTATTAATAAACCTCAAGAAAACGTTTAGAAGATCGTTGAAGCCATCAAA
TCTGAAGCAGAAGAGAAGGCTGAACAAATTAAAAAGAATGCTGAAGAATAATTTAAGATT
CAAAAAAATAATATTGTCAATACTGAAAAGGATAAAATCATTGAAGAATATAAAAAGAGA
TTAGAAAAGCTCATAGTTGACAGAAGAATGTAATAATTTTATTGAAAAAGAATTGAAAAT
AATTATATTTTTAAGCAGTAAATTTAACAATTAAGCACATTTATTACTAAAAAAATCTAA
AATTATTTTCAAATATCTTAAAATAGTCAACGTTCTGCCAAAATCAATGAATAGAGATTA
GAAAAAATGAAAGCTCGTTTTGACTTTATTGAAAAGCTAAAGGGTGAAATTTCTAATAAA
ATCGTTTAATCCGTTTCAGACCCTAATAAATATAAAAATGTCTTCAAGCAACTTATAATC
TAGGTAATCCATTCAGCTAATATCCTTCATGTTTTAACTAAAAAATATAATTGCATTTGA
AATAAAATAATCAAATGATAAATATAGCAGCAGATAAAATAGTAATCCTATAATTTAGAT
TAGCATTAACAAAGACCAATAAGGTTGTTCAAAAAATATTAAAATGTTAGCATTGAAATT
TAGGTTAAAAATTTAGCATAATTGAATAAAAAATAAAATGATTCTATGGCAAATATTATA
TATCGAAATAAATATCAAACTTTGCTAAACACAAGATATATTAATTACTGCTTTTAATTA
AGTTATTAGCAATAAAAATCGTCATAGTTAGTGCATAAAAGGATTCAACAAATGCAAGTG
AATCAAATATTTAAGATCGATTTTTATAGTGATAAAAAAGTAGAATTATATAAATTTTTA
TTGTATTATTAGGCCTTGATCAAGTTGATGGAACCTAAGGTTGAGTTAAAAGTAATGAAG
AAAGATTTGTAGCTTGCTCGTGAAGTAAAGACAGAATGTGAAAATGAATTCAAGGCAATT
GCTAAGAGAGAATGTAATAGAGACTTTAACTGTACTATTATTATAAATGAATACCACTCT
CTTGAAGAAGAAAATCCTAAAGTGTAAACAAATTCATCCAAAGCATTTACTTTATCAATA
TTATTACTATTATTTATTTAGTATTGGAGGTATTGTTCTCACTTGCGATGGTGGTAGAAT
TTAGGTAAACAACACTTTAAATGCAAGAGTTGATCTTGCTTTCTAAGAATTCCTTCCTGA
CATCAGAAGAATTCTTTTCCCTTCTATGAAATGA>TTHERM_00193660(protein)
MADFINKPQENVQKIVEAIKSEAEEKAEQIKKNAEEQFKIQKNNIVNTEKDKIIEEYKKR
LEKLIVDRRIQRSAKINEQRLEKMKARFDFIEKLKGEISNKIVQSVSDPNKYKNVFKQLI
IQALIKLMEPKVELKVMKKDLQLAREVKTECENEFKAIAKRECNRDFNCTIIINEYHSLE
EENPKVIGGIVLTCDGGRIQVNNTLNARVDLAFQEFLPDIRRILFPSMK