Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_01079200Standard Name
HTA4 (Histone Two A 4)Aliases
PreTt17435 | 221.m00055 | 3832.m00156 | HTAYDescription
HTAY histone H2A; Core histone H2A variantGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Molecular Function
- DNA binding (IEA) | GO:0003677
- protein binding (IEA) | GO:0005515
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
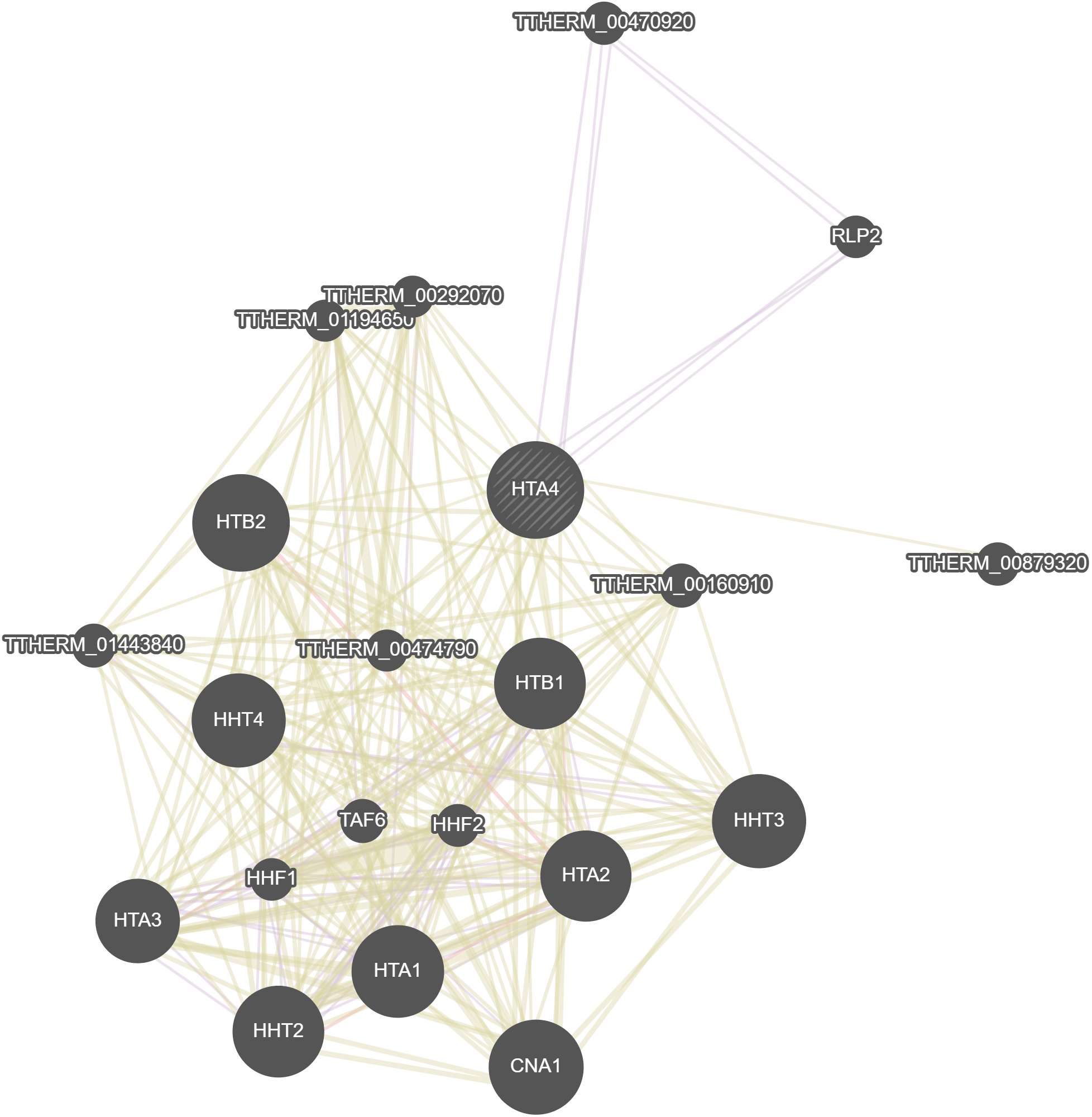
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
- ( SD01769 ) Macronucleus: Flag C-terminal tagged HTAY gene fragment coding for AA123 to 388 into MTT1 locus
- ( SD01786 ) Macronucleus: Neo3 KO of HTAY(may be incomplete) and HTAY with two HA tags on the N-terminus inserted into MTT1 locus with neo2 cassete in 5' flanking region
- ( SD01997 ) Macronucleus: Neo3 KO of HTAY (may be incomplete) and HTAY with two HA tags on the N-terminus inserted into MTT1 locus with neo2 cassete in 5' flanking region
- ( SD02200 ) Macronucleus: Neo3 into HTAY, Chimeric YN-HTAX into MTT1 locus
- ( SD02201 ) Macronucleus: Neo3 into HTAY, Chimeric YN-HTAX into MTT1 locus
- ( SD02202 ) Macronucleus: Neo3 into HTAY, Chimeric YN-HTAX into MTT1 locus
- ( SD02203 ) Macronucleus: Neo3 into HTAY, Chimeric YN-HTAX into MTT1 locus
- ( SD02368 ) Macronucleus: 2 HA tagged HTAY into MTT1 with neo2 in 5' flank
- ( SD02381 ) Micronucleus: Neo3 replaces bases coding for AA258-388 Macronucleus: Neo3 replaces bases coding for AA258-388
- ( SD02676 ) Micronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene Macronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene
- ( SD02677 ) Micronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene Macronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene
- ( SD02678 ) Micronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene
- ( SD02679 ) Micronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene
- ( SD02680 ) Micronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene
- ( SD02700 ) Macronucleus: 3HA n-terminal tagged HTAY gene fragment coding for AA123 to 388into MTT1 locus
- ( SD02701 ) Macronucleus: Flag tagged 2/3 c-terminus of HTAY into MTT1 locus
- ( SD02808 ) Macronucleus: Partial ko of HTAY coding with neo3
- ( SD02809 ) Macronucleus: Partial ko of HTAY coding with neo3
- ( SD02881 ) Macronucleus: Partial replacement of MTT1 coding by N-terminus of HTAY, aa 1-122, with neo2 in 5’ flank of MTT1
- ( SD02882 ) Macronucleus: Region of HTAY gene encoding AA 123 to 257 with two HA tags at n-terminus was inserted into the MTT1 gene.
- ( SD02883 ) Macronucleus: Region of HTAY gene encoding AA 258 to 388 with two HA tags at n-terminus was inserted into the MTT1 gene.
- ( SD02884 ) Macronucleus: Region of HTAY gene encoding AA 1 to 258 with two HA tags at n-terminus was inserted into the MTT1 gene.
- ( SD02885 ) Macronucleus: Region of HTAY gene encoding AA 1 to 122 and 258 to 388 with two HA tags at n-terminus was inserted into the MTT1 gene.
 Homologs
Homologs
Source Identifier Score Tetrahymena borealis EI9_20318.1 9.997093137033517e-262 Description: core histone H2A/H2B/H3/H4 family protein (381 aa) Oxytricha Contig9320.0.g50 2.000130478826482e-26 Description: Core histone H2A/H2B/H3/H4 SGD YDR225W 4.0013320766793207e-25 Description: HTA1 Histone H2A, core histone protein required for chromatin assembly and chromosome function; one of two nearly identical subtypes (see also HTA2); DNA damage-dependent phosphorylation by Mec1p facilitates DNA repair; acetylated by Nat4p WormBase WBGene00001886 2.000620012690291e-23 Description: locus:his-12 Histone H2A status:Predicted UniProt:P09588 protein_id:CAB05838.1 DictyBase DDB_G0279667 4.0002685161515803e-16 Description: H2AX on chromosome: 3 position 2410613 to 2411077 Stentor Coeruleus SteCoe_25962 0.0000000000018795288165390832 Description: None
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:22426537: Song X, Bowen J, Miao W, Liu Y, Gorovsky MA (2012) The nonhistone, N-terminal tail of an essential, chimeric H2A variant regulates mitotic H3-S10 dephosphorylation. Genes & development 26(6):615-29
- Ref:16933976: Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA,
 Sequences
Sequences
>TTHERM_01079200(coding)
ATGTTGATTAACTCTATATTAGATTTTAATAGTATCAAATCTTTTAATTCGATAGTATAA
ATAAAAAACTTATATATACAAATAAAGATATTAATAATAACTTTGACAAACATTTTTACA
AAAAATAAAAAATAAGTTAAATAAACACAACAATAATCTTTAAAAATGTCAACTGACTAA
GAAAAGATAATAAAATAAGTATTAGATTCAATAGCAGAGCAATAGGGAGATAGTAGAGAT
GATGAAGTTTATGTGATAGAAATAATGTCATTACGCTTTGAGAAATTTGATAATCGAATT
AAGTAGAAGATTGAAAGATTTACATCTCTTTAAATGCTAACAATCAACGATTGCCTTATC
TCTGATTTAACTAACTTCCCTCATGTTCCTTCTTTGATTAGATTAGACTTGGTCTTTAAT
AAAATTACTGGAGACTAGCTTTAATATTTGAGAGGATCAAGACATTTACAAACTTTAATG
CTTGGTGCAAATTAAATTGAAGAGATCGAAGACTTGAAGAGACTCGGTTAGATGAGAGAA
TTAATTCAACTAGATTTATTGAATAATCCAGTTGTTAATACAAATAATTATAGAAATTTA
GTTTTTAATTTGTTTCCTTCTTTGGTAATATTAGACACTTTAGATAAAAATGGAATTGAT
TAAGAAAAAGCAGCTCTTGATATTTCTGCTTCAAGAGTGCCTGATAACTTATTTGATAAA
TCTAAGCCTGTATAAAATGTTTCCAAATAAGTAGTTAAAAATGCTAAAGCTACTTAAAAT
AATCTTTTCAGTTCTACACAAAAAGTAGCTTAAGTTAAATAAATTCCCAAAATTGTACCT
GCAAAAGTATCTAAGCCTTCTCAAGCATCAGTTTAAGACAACAAATCTAATGTAGTTTAA
GCAAAAGTTACTGCTGCTGTAAGTGTTGGTAGAAAAGCACCTGCATCAAGAAATGGTGGA
GTACCATCAAAAGCTGGAAAAGGCAAAATGAATGCTTTTTCAAAGTAGTCTACATCTCAA
AAAAGTGGTTTAGTATTTCCTTGTGGAAGACTTAGAAGATACTTGAAACAGGCTTGCAAA
CAATTTAGAGTTTCTTCAAGCTGTAACATTTACTTAGCTGGTGTTCTTGAATACTTAGCA
GCAGAAGTCTTAGAAACTGCAGGAAATATAGCAAAAAACAATAGGCTAAGTAGAATTAAC
CCAGTTCATATAAGAGAAGCTTTTAGAAACGATGCTGAGTTAAACTAATTTGTAAATGGT
ACTATTATAGCTGAAGGAGGAAGCACAAGTACTAATTTTGTCCTTCCAATTATTAATAAA
TCCAAAAAATGA>TTHERM_01079200(gene)
ATGTTGATTAACTCTATATTAGATTTTAATAGTATCAAATCTTTTAATTCGATAGTATAA
ATAAAAAACTTATATATACAAATAAAGATATTAATAATAACTTTGACAAACATTTTTACA
AAAAATAAAAAATAAGTTAAATAAACACAACAATAATCTTTAAAAATGTCAACTGACTAA
GAAAAGATAATAAAATAAGTATTAGATTCAATAGCAGAGCAATAGGGAGATAGTAGAGAT
GATGAAGTTTATGTGATAGAAATAATGTCATTACGCTTTGAGAAATTTGATAATCGAATT
AAGTAGAAGATTGAAAGATTTACATCTCTTTAAATGCTAACAATCAACGATTGCCTTATC
TCTGATTTAACTAACTTCCCTCATGTTCCTTCTTTGATTAGATTAGACTTGGTCTTTAAT
AAAATTACTGGAGACTAGCTTTAATATTTGAGAGGATCAAGACATTTACAAACTTTAATG
CTTGGTGCAAATTAAATTGAAGAGATCGAAGACTTGAAGAGACTCGGTTAGATGAGAGAA
TTAATTCAACTAGATTTATTGAATAATCCAGTTGTTAATACAAATAATTATAGAAATTTA
GTTTTTAATTTGTTTCCTTCTTTGGTAATATTAGACACTTTAGATAAAAATGGAATTGAT
TAAGAAAAAGCAGCTCTTGATATTTCTGCTTCAAGAGTGCCTGATAACTTATTTGATAAA
TCTAAGCCTGTATAAAATGTTTCCAAATAAGTAGTTAAAAATGCTAAAGCTACTTAAAAT
AATCTTTTCAGTTCTACACAAAAAGTAGCTTAAGTTAAATAAATTCCCAAAATTGTACCT
GCAAAAGTATCTAAGCCTTCTCAAGCATCAGTTTAAGACAACAAATCTAATGTAGTTTAA
GCAAAAGTTACTGCTGCTGTAAGTGTTGGTAGAAAAGCACCTGCATCAAGAAATGGTGGA
GTACCATCAAAAGCTGGAAAAGGCAAAATGAATGCTTTTTCAAAGTAGTCTACATCTCAA
AAAAGTGGTTTAGTATTTCCTTGTGGAAGACTTAGAAGATACTTGAAACAGGCTTGCAAA
CAATTTAGAGTTTCTTCAAGCTGTAACATTTACTTAGCTGGTGTTCTTGAATACTTAGCA
GCAGAAGTCTTAGAAACTGCAGGAAATATAGCAAAAAACAATAGGCTAAGTAGAATTAAC
CCAGTTCATATAAGAGAAGCTTTTAGAAACGATGCTGAGTTAAACTAATTTGTAAATGGT
ACTATTATAGCTGAAGGAGGAAGCACAAGTACTAATTTTGTCCTTCCAATTATTAATAAA
TCCAAAAAATGA>TTHERM_01079200(protein)
MLINSILDFNSIKSFNSIVQIKNLYIQIKILIITLTNIFTKNKKQVKQTQQQSLKMSTDQ
EKIIKQVLDSIAEQQGDSRDDEVYVIEIMSLRFEKFDNRIKQKIERFTSLQMLTINDCLI
SDLTNFPHVPSLIRLDLVFNKITGDQLQYLRGSRHLQTLMLGANQIEEIEDLKRLGQMRE
LIQLDLLNNPVVNTNNYRNLVFNLFPSLVILDTLDKNGIDQEKAALDISASRVPDNLFDK
SKPVQNVSKQVVKNAKATQNNLFSSTQKVAQVKQIPKIVPAKVSKPSQASVQDNKSNVVQ
AKVTAAVSVGRKAPASRNGGVPSKAGKGKMNAFSKQSTSQKSGLVFPCGRLRRYLKQACK
QFRVSSSCNIYLAGVLEYLAAEVLETAGNIAKNNRLSRINPVHIREAFRNDAELNQFVNG
TIIAEGGSTSTNFVLPIINKSKK