Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00486480Standard Name
ENO1 (ENOlase 1)Aliases
PreTt19579 | 59.m00217 | 3680.m00074Description
ENO1 carboxy-terminal TIM barrel domain enolase; Enolase; portions of this gene may have been laterally transferredGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- cilium (IDA) | GO:0005929
- microtubule basal body (IDA) | GO:0005932
- phosphopyruvate hydratase complex (IEA) | GO:0000015
Molecular Function
- 5'-3' RNA exonuclease activity (ISS) | GO:0004534
- magnesium ion binding (IEA) | GO:0000287
- phosphopyruvate hydratase activity (IEA) | GO:0004634
Biological Process
- glycolytic process (IEA) | GO:0006096
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
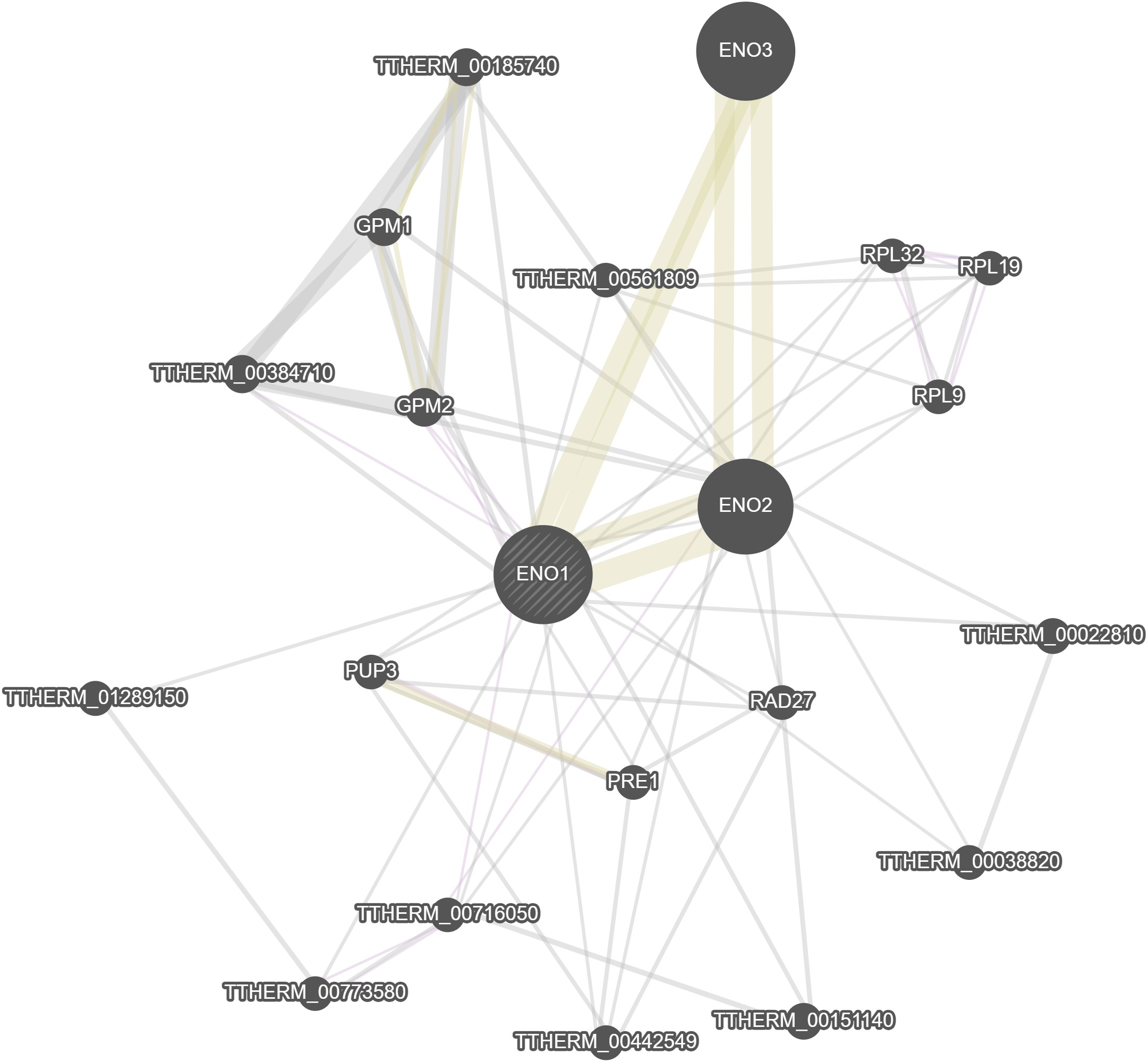
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
Source Identifier Score Stentor Coeruleus SteCoe_4496 0 Description: None Tetrahymena borealis EI9_12148.1 9.997093137033517e-262 Description: enolase (441 aa) DictyBase DDB_G0283137 9.997093137033517e-262 Description: enoA on chromosome: 4 position 225309 to 226956 Oxytricha Contig15891.0.g72 9.997093137033517e-262 Description: Enolase, C-terminal TIM barrel domain Tetrahymena borealis EI9_12253.1 9.997093137033517e-262 Description: GHMP kinase ATP-binding protein (423 aa) WormBase WBGene00011884 1.9999987447608204e-178 Description: locus:enol-1 Enolase status:Confirmed UniProt:Q27527 protein_id:CAA92692.1 Tetrahymena borealis EI9_01117.1 1.9999987447608204e-178 Description: tubulin-tyrosine ligase (671 aa) SGD YGR254W 2.0002969069156684e-168 Description: ENO1 Enolase I, a phosphopyruvate hydratase that catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate during glycolysis and the reverse reaction during gluconeogenesis; expression is repressed in response to glucose
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:17785518: Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Culver BP, Yates JR, Winey M (2007) New Tetrahymena basal body protein components identify basal body domain structure. The Journal of cell biology 178(6):905-12
- Ref:11526220: Keeling PJ and Palmer JD (2001) Lateral transfer at the gene and subgenic levels in the evolution of eukaryotic enolase. Proc Natl Acad Sci U S A 98(19):10745-50
 Sequences
Sequences
>TTHERM_00486480(coding)
ATGGCTACTATTAAAGATATCAAAGCTAGAGAAATACTAGATTCTAGAGGTAATCCTACT
GTTGAAGTTGATTTGACAGTAGATAATGGTCAAGTTTTTAGAGCTGCTGTTCCTTCTGGT
GCTTCAACAGGTATCTATGAAGCCTGTGAACTTAGAGACGGGGATAATCACAGATATTTA
GGGAAAGGTGTTTTGAAGGCTGTAAACAATGTGAACACCATCATTAAACCTCATTTAATT
GGTAAAAATGTTACTGAATAAGAATAATTAGATAAGTTAATGGTAGAATAGCTTGATGGA
ACTAAAAATGAATGGGGTTGGTGCAAAAGCAAATTAGGAGCAAATGCTATTTTAGCAGTT
TCTATGGCTGTTGCCAGAGCTGCCGCAGCTCATAAGAACCTTCCTTTATACAAATACTTG
TCTCATTTGAATGGATAAGACAACCTAACCCAATTCAATTTACCTCTTCCATTCTTCAAT
GTCATTAATGGTGGAAAGCATGCAGGAAACAAATTAGCTTTCTAAGAGTTTATGTTCACA
CCATGCGGTGCATCTAACTTTGCAGAAGGTCTAAGAATGGGTGCTGAGGTTTATCACAAC
TTAAAGAAAGTAATTGAAAAAAAGTACGGACAAACAGCTACTAATGTAGGTGATGAAGGA
GGTTTTGCCCCAGATACTAGTAATCCAGATGAAGCATTAGAATTACTAGTTGAAGCTATC
AAGAAGGCAGGTCATGAAGGAAAGATTAAAATATCTATGGATGTTGCAGCTTCTGAATTC
TACGACGCTAAAACTAAGCTTTATGATTTAGATTTTAAGAATAGTAACTCAGATGGTAAG
CATAAGTTAACTTCTGAATAGTTAGTTGAATACTTTGCCAATTTAGTTTCAAAATATCCA
ATTTTTTCAATTGAAGATCCTTTCGACTAGGATGACTGGGCTGCTTATGCTCAATTGACT
GCTCGTATTGGTAATTAAGTCAAGATTGTTGGTGATGACTTACTTGTTACTAACCCTTTA
CGTGCAGAAAAAGCTATTTAAGAAAAAGCCTGTAATTATCTTCTACTTAAAGTCAATCAA
ATTGGAACAGTAACAGAAGCAATTTAAACATATCAAATTGTCTCTAAAGCCAACTTTGGA
ACTATGGTCTCACATAGATCAGGTGAAACTGAAGATACTTTCATAGCTGATTTAGTTGTA
GGAATAGGTGCTGCTACAATTAAAACTGGTGCTCCCTGCAGATCTGAAAGATTAGCTAAA
TATAATTAGCTTATCAGAATTGAAGAAGAGCTTGGTCAAAATTCTACTTATGGAAAAAAT
CACTGA>TTHERM_00486480(gene)
ATGGCTACTATTAAAGATATCAAAGCTAGAGAAATACTAGATTCTAGAGGTAATCCTACT
GTTGAAGTTGATTTGACAGTAGATAATGGTCAAGTTTTTAGAGCTGCTGTTCCTTCTGGT
GCTTCAACAGGTATCTATGAAGCCTGTGAACTTAGAGACGGGGATAATCACAGATATTTA
GGGAAAGGTGTTTTGAAGGCTGTAAACAATGTGAACACCATCATTAAACCTCATTTAATT
GGTAAAAATGTTACTGAATAAGAATAATTAGATAAGTTAATGGTAGAATAGCTTGATGGA
ACTAAAAATGAATGGGGTTGGTGCAAAAGCAAATTAGGAGCAAATGCTATTTTAGCAGTT
TCTATGGCTGTTGCCAGAGCTGCCGCAGCTCATAAGAACCTTCCTTTATACAAGTAAATA
TTTTTAAAATTCTATTTCAAATTATAAAAAGCCTCTTTACTTTATTTAAAAACCAAATCT
TATCATCTTTGCATTAACATTTTTTATGGAAGATTTTAAAACTTTGCAGCAATAAATACT
TAGGTTAAAAGCTTCATATAAACAAAAAAACATGAAATTATTTATTCCTAACTTAATATA
AATAAAATTAAACTAGATACTTGTCTCATTTGAATGGATAAGACAACCTAACCCAATTCA
ATTTACCTCTTCCATTCTTCAATGTCATTAATGGTGGAAAGCATGCAGGAAACAAATTAG
CTTTCTAAGAGTTTATGTTCACACCATGCGGTGCATCTAACTTTGCAGAAGGTCTAAGAA
TGGGTGCTGAGGTTTATCACAACTTAAAGAAAGTAATTGAAAAAAAGTACGGACAAACAG
CTACTAATGTAGGTGATGAAGGAGGTTTTGCCCCAGATACTAGTAATCCAGATGAAGCAT
TAGAATTACTAGTTGAAGCTATCAAGAAGGCAGGTCATGAAGGAAAGATTAAAATATCTA
TGGATGTTGCAGCTTCTGAATTCTACGACGCTAAAACTAAGCTTTATGATTTAGATTTTA
AGAATAGTAACTCAGATGGTAAGCATAAGTTAACTTCTGAATAGTTAGTTGAATACTTTG
CCAATTTAGTTTCAAAATATCCAATTTTTTCAATTGAAGATCCTTTCGACTAGGATGACT
GGGCTGCTTATGCTCAATTGACTGCTCGTATTGGTAATTAAGTCAAGATTGTTGGTGATG
ACTTACTTGTTACTAACCCTTTACGTGCAGAAAAAGCTATTTAAGAAAAAGCCTGTAATT
ATCTTCTACTTAAAGTCAATCAAATTGGAACAGTAACAGAAGCAATTTAAACATATCAAA
TTGTCTCTAAAGCCAACTTTGGAACTATGGTCTCACATAGATCAGGTGAAACTGAAGATA
CTTTCATAGCTGATTTAGTTGTAGGAATAGGTGCTGCTACAATTAAAACTGGTGCTCCCT
GCAGATCTGAAAGATTAGCTAAATATAATTAGCTTATCAGAATTGAAGAAGAGCTTGGTC
AAAATTCTACTTATGGAAAAAATCACTGA>TTHERM_00486480(protein)
MATIKDIKAREILDSRGNPTVEVDLTVDNGQVFRAAVPSGASTGIYEACELRDGDNHRYL
GKGVLKAVNNVNTIIKPHLIGKNVTEQEQLDKLMVEQLDGTKNEWGWCKSKLGANAILAV
SMAVARAAAAHKNLPLYKYLSHLNGQDNLTQFNLPLPFFNVINGGKHAGNKLAFQEFMFT
PCGASNFAEGLRMGAEVYHNLKKVIEKKYGQTATNVGDEGGFAPDTSNPDEALELLVEAI
KKAGHEGKIKISMDVAASEFYDAKTKLYDLDFKNSNSDGKHKLTSEQLVEYFANLVSKYP
IFSIEDPFDQDDWAAYAQLTARIGNQVKIVGDDLLVTNPLRAEKAIQEKACNYLLLKVNQ
IGTVTEAIQTYQIVSKANFGTMVSHRSGETEDTFIADLVVGIGAATIKTGAPCRSERLAK
YNQLIRIEEELGQNSTYGKNH