Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00516380Standard Name
CCF6 (C-terminal Crystallin Fold 6)Aliases
PreTt10730 | 64.m00210 | 3723.m00066Description
CCF6 carboxy-terminal crystallin fold protein 7p putative; C-terminal crystallin fold protein; domain structure suggests localization in dense core granules; contains signal peptide and additional 200-residue motif weakly conserved among CCF6- CCF7- CCF8- and IGR1Genome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- dense core granule (ISS) | GO:0031045
 Domains
Domains
No Data fetched for Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
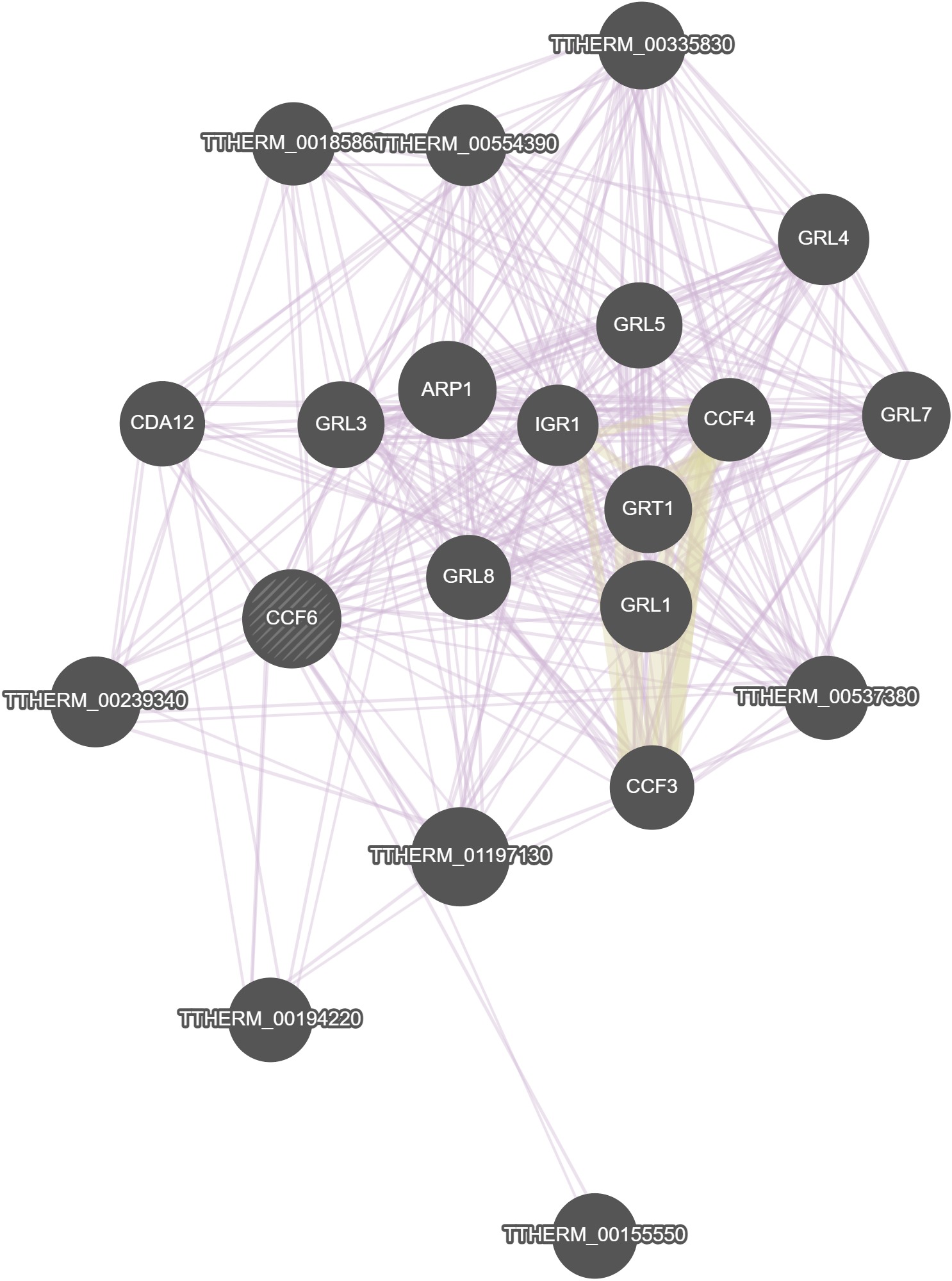
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
Source Identifier Score Tetrahymena borealis EI9_15450.1 4.001282277195165e-160 Description: hypothetical protein (310 aa) Tetrahymena borealis EI9_06070.1 4.001282277195165e-160 Description: hypothetical protein (1190 aa)
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:16014006: Bowman GR, Smith DG, Michael Siu KW, Pearlman RE, Turkewitz AP (2005) Genomic and proteomic evidence for a second family of dense core granule cargo proteins in Tetrahymena thermophila. The Journal of eukaryotic microbiology 52(4):291-7
 Sequences
Sequences
>TTHERM_00516380(coding)
ATGCTTCTGTTTTATACTATTATTAAAGCAGTAATTAAGCATATGAAAGTTTTTGATTTT
TTGATATTTAATTTGATGAGATTAGTGCTAAAATCAGATAATAAATTAAAAATATAATCT
TATATTGATATAGATAAAGATAAATTGTTTAGCAAAAAAATAATGAGAAAGGCTAATTTA
ATATTTGTTATCTTAGGATTGACCCTAAGCGCCTGCTACGCTTAAACAGTAGTTGTCACT
GATTTTACTATAGCTGCTGGTAACTCAGTACCCTATATTAAAACATTCGAAAACTAATGG
AACGATGAATTAGAATTCGCTGTTTATGGATGGATCAGAATACCAAGTTGGGTTAATAGT
GAAGTTTAAGTATTCCGTTTATCTGCTAACAAAGACATAAATTCTAATACTCTTGGTGAT
AGAGTTTTGAAGTCTTTTGTCCAAGATAACAAAGTTATGTTCGAGACTTATGATGCTTAG
AATTACGACCCTAAAGTTTAACATTCTTATTTGAACCAAGGTGATAGTTTTGGATAATGG
TTCTTCATTTACTAGGGCTACAATCCTAAAACAACAAAGACTTATGGTTGGGTTTAAAAC
GCAGCTGGAAAAGGTTAAGGAGGTTACAGCAATTAATTAGCTAGACATACCAAGTCTAAT
TTCTATTAAGTAATCGTAGGTCCTAATTCAGGCCTTAAAGGAAGTGATAAAGTAGAAGTC
AAAATTCTTTGGATTTAAGCTGGATCTGGTGCTTACAGAGAAAACAACTTTGATACAAGA
GATACCAATGTAGTAGTAGTACCTATCCCATCTGTTAAATGCCCTACAGTTTACTCTCAA
TGTGATTACAAGGGAGATTCATCATCTTACTGTTAAAGTGTCCCTTCTGTCAAAATCCCT
GTAATTAAGAGTGTTGTAATCCCTGATGGATTCAATAAGATTACTGTGTATAATCTTGAA
AATTACAAAGGTAAGACCGTTATTTTTAACAAAAGCGTTTCTTGTCTCGACAAATTTGAA
TTTGCCTTCCTAGAGAGAGAAGGATATATCACAGTTATTGATAATAAAGAGTCAAAAAAA
TCTAAAAATGCTCTTAGATCAAACAGAAAATGA>TTHERM_00516380(gene)
ATGCTTCTGTTTTATACTATTATTAAAGCAGTAATTAAGCATATGAAAGTTTTTGATTTT
TTGATATTTAATTTGATGAGATTAGTGCTAAAATCAGATAATAAATTAAAAATATAATCT
TATATTGATATAGATAAAGATAAATTGTTTAGCAAAAAAATAATGAGAAAGGCTAATTTA
ATATTTGTTATCTTAGGATTGACCCTAAGCGCCTGCTACGCTTAAACAGTAGTTGTCACT
GATTTTACTATAGCTGCTGGTAACTCAGTACCCTATATTAAAACATTCGAAAACTAATGG
AACGATGAATTAGAATTCGCTGTTTATGGATGGATCAGAATACCAAGTTGGGTTAATAGT
GAAGTTTAAGTATTCCGTTTATCTGCTAACAAAGACATAAATTCTAATACTCTTGGTGAT
AGAGTTTTGAAGTCTTTTGTCCAAGATAACAAAGTTATGTTCGAGACTTATGATGCTTAG
AATTACGACCCTAAAGTTTAACATTCTTATTTGAACCAAGGTGATAGTTTTGGATAATGG
TTCTTCATTTACTAGGGCTACAATCCTAAAACAACAAAGACTTATGGTTGGGTTTAAAAC
GCAGCTGGAAAAGGTTAAGGAGGTTACAGCAATTAATTAGCTAGACATACCAAGTCTAAT
TTCTATTAAGTAATCGTAGGTCCTAATTCAGGCCTTAAAGGAAGTGATAAAGTAGAAGTC
AAAATTCTTTGGATTTAAGCTGGATCTGGTGCTTACAGAGAAAACAACTTTGATACAAGA
GATACCAATGTAGTAGTAGTACCTATCCCATCTGTTAAATGCCCTACAGTTTACTCTCAA
TGTGATTACAAGGGAGATTCATCATCTTACTGTTAAAGTGTCCCTTCTGTCAAAATCCCT
GTAATTAAGAGTGTTGTAATCCCTGATGGATTCAATAAGATTACTGTGTATAATCTTGAA
AATTACAAAGGTAAGACCGTTATTTTTAACAAAAGCGTTTCTTGTCTCGACAAATTTGAA
TTTGCCTTCCTAGAGAGAGAAGGATATATCACAGTTATTGATAATAAAGAGTCAAAAAAA
TCTAAAAATGCTCTTAGATCAAACAGAAAATGA>TTHERM_00516380(protein)
MLLFYTIIKAVIKHMKVFDFLIFNLMRLVLKSDNKLKIQSYIDIDKDKLFSKKIMRKANL
IFVILGLTLSACYAQTVVVTDFTIAAGNSVPYIKTFENQWNDELEFAVYGWIRIPSWVNS
EVQVFRLSANKDINSNTLGDRVLKSFVQDNKVMFETYDAQNYDPKVQHSYLNQGDSFGQW
FFIYQGYNPKTTKTYGWVQNAAGKGQGGYSNQLARHTKSNFYQVIVGPNSGLKGSDKVEV
KILWIQAGSGAYRENNFDTRDTNVVVVPIPSVKCPTVYSQCDYKGDSSSYCQSVPSVKIP
VIKSVVIPDGFNKITVYNLENYKGKTVIFNKSVSCLDKFEFAFLEREGYITVIDNKESKK
SKNALRSNRK