Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00570550Standard Name
CCF13 (C-terminal Crystallin Fold 13 )Aliases
PreTt10484 | 76.m00186 | 10484 | 3810.m02046Description
CCF13 carboxy-terminal crystallin fold protein; C-terminal crystallin fold protein; domain structure suggests localization in dense core granulesGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- dense core granule (ISS) | GO:0031045
 Domains
Domains
No Data fetched for Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
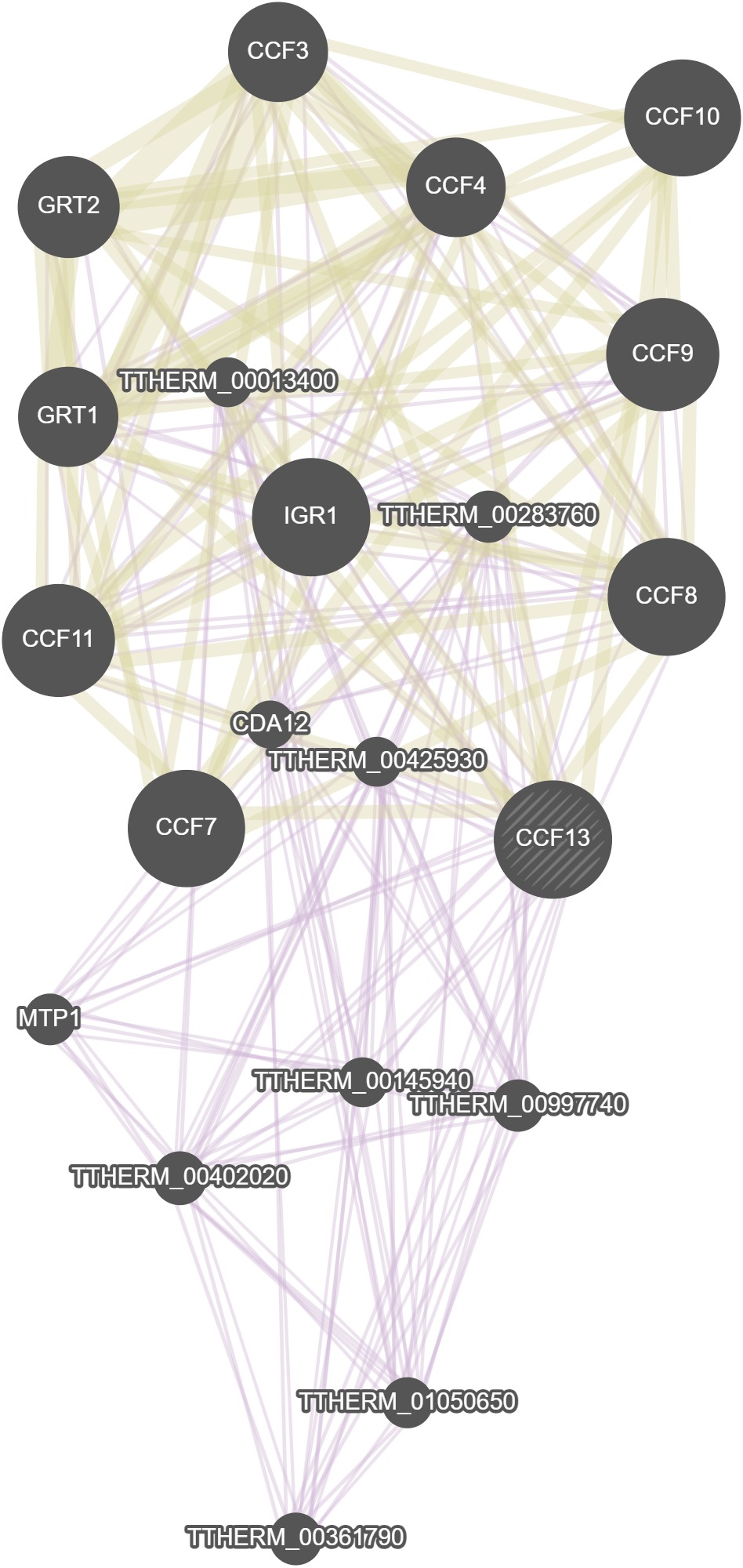
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
Source Identifier Score Tetrahymena borealis EI9_03454.1 2.000130478826482e-26 Description: hypothetical protein (185 aa) Tetrahymena borealis EI9_19845.1 2.000130478826482e-26 Description: hypothetical protein (1107 aa)
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00570550(coding)
ATGAAAACTACTTTGGTGGTATTTATTATTTTAGGATTAGCCTTTTCTAAGATAGGCGCA
AAATAAAGTTAAACATAGAGTGTTACAATCAACAATAATAATCCAATGTAGAGTGTTACG
ATCACCAAAGGAGGGTAGCCATAAGGTGATACTACTACATGGGATGGAGTTTATAAAAGT
TTGAAAGTGACTGATATTGGTTAAAAACCTGTAGATCCTGTTGTTACTCCTGCTCAATGG
GATGGAGTTTATAAGAGTTTGAAAGTGACTAATGATGTTACAAAACCTGAAGAACCTCCT
GTTTTACCACCAAAAGTATGCCCCACTCTTTACAAGCAATGCTATTTCAAAGGAGATCCA
ATCTAGCTTTGTGATAAAATTCCTAAATTCGATATTAAAACCATTCATAGTGTCTGGATC
CCTGATGGCAGCAGAGTTACTCTTTATGAAAAAAGCGACTATAGCGGAAAATCAATAACC
TTCAAAGAGACTATTCCTTGCATTGATTCATTAAGTTTCTCTTGA>TTHERM_00570550(gene)
ATGAAAACTACTTTGGTGGTATTTATTATTTTAGGATTAGCCTTTTCTAAGATAGGCGCA
AAATAAAGTTAAACATAGAGTGTTACAATCAACAATAATAATCCAATGTAGAGTGTTACG
ATCACCAAAGGAGGGTAGCCATAAGGTGATACTACTACATGGGATGGAGTTTATAAAAGT
TTGAAAGTGACTGATATTGGTTAAAAACCTGTAGATCCTGTTGTTACTCCTGCTCAATGG
GATGGAGTTTATAAGAGTTTGAAAGTGACTAATGATGTTACAAAACCTGAAGAACCTCCT
GTTTTACCACCAAAAGTATGCCCCACTCTTTACAAGCAATGCTATTTCAAAGGAGATCCA
ATCTAGCTTTGTGATAAAATTCCTAAATTCGATATTAAAACCATTCATAGTGTCTGGATC
CCTGATGGCAGCAGAGTTACTCTTTATGAAAAAAGCGACTATAGCGGAAAATCAATAACC
TTCAAAGAGACTATTCCTTGCATTGATTCATTAAGTTTCTCTTGA>TTHERM_00570550(protein)
MKTTLVVFIILGLAFSKIGAKQSQTQSVTINNNNPMQSVTITKGGQPQGDTTTWDGVYKS
LKVTDIGQKPVDPVVTPAQWDGVYKSLKVTNDVTKPEEPPVLPPKVCPTLYKQCYFKGDP
IQLCDKIPKFDIKTIHSVWIPDGSRVTLYEKSDYSGKSITFKETIPCIDSLSFS