Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00675850Standard Name
LIA3 (localized in macronuclear anlagen 3)Aliases
PreTt11026 | 101.m00181Description
LIA3 a parallel G quadruplex-binding protein that regulates DNA elimination boundaries; hypothetical proteinGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
No Data fetched for Gene Ontology Annotations
 Domains
Domains
No Data fetched for Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
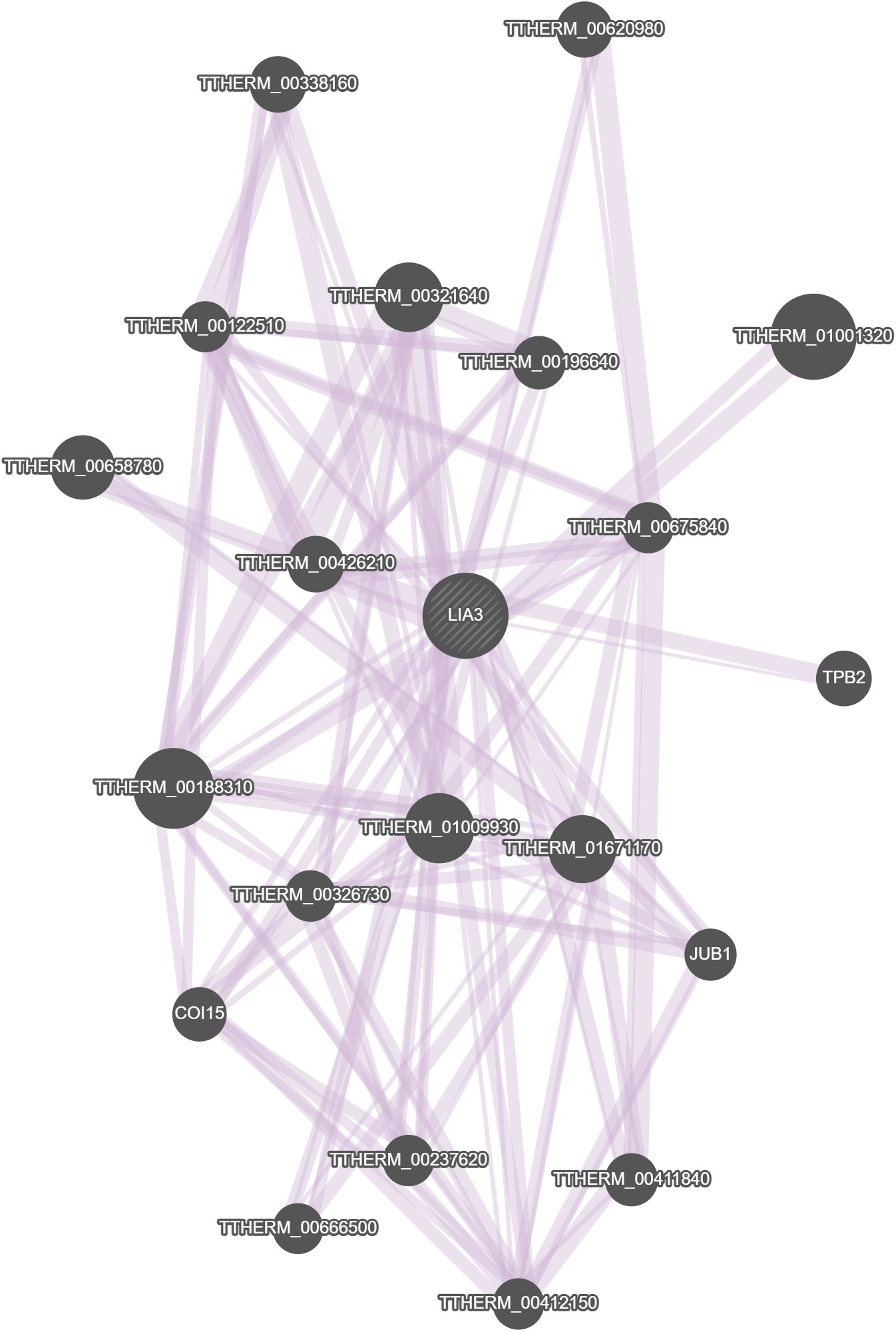
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No. Gene Name(s) Paragraph Text 2318 LIA3 In cells lacking LIA3 (ΔLIA3), the excision of IESs bounded by specific G-rich polypurine tracts was impaired and imprecise, whereas the removal of IESs without such controlling sequences was unaffected. Lia3p binds to oligonucleotides containing these polypurine tracts, which form parallel G-quadruplex structures in vitro.
 Associated Literature
Associated Literature
- Ref:30918956: Lin CG, Chao JL, Tsai HK, Chalker D, Yao MC (2019) Setting boundaries for genome-wide heterochromatic DNA deletions through flanking inverted repeats in Tetrahymena thermophila. Nucleic acids research 47(10):5181-5192
- Ref:31194876: Jaspan VN, Taye ME, Carle CM, Chung JJ, Chalker DL (2019) Boundaries of eliminated heterochromatin of Tetrahymena are positioned by the DNA-binding protein Ltl1. Nucleic acids research ( ):
- Ref:26950070: Carle CM, Zaher HS, Chalker DL (2016) A Parallel G Quadruplex-Binding Protein Regulates the Boundaries of DNA Elimination Events of Tetrahymena thermophila. PLoS genetics 12(3):e1005842
- Ref:17519286: Yao MC, Yao CH, Halasz LM, Fuller P, Rexer CH, Wang SH, Jain R, Coyne RS, Chalker DL (2007) Identification of novel chromatin-associated proteins involved in programmed genome rearrangements in Tetrahymena. Journal of cell science 120(Pt 12):1978-89
 Sequences
Sequences
>TTHERM_00675850(coding)
ATGAAATCTCATTCAAAGAAATACCAAGTAACAAATAATTTAGATGCTGACAATGAATTA
TATCCTCAATCACAAATTTAGAGTTCCAATAATGATTAGTAAACTCAACTGAACAGTCAA
GAAGAAAATAACCCTGATATGATTGAAACTGAAGATATAAACTTTAATCCAAATAACTCC
AATAAACCAACAAATAGTTCTTATAAAATTCAAACAATAAACTTAGAGAATAACAATCTA
GGTGAAAATACTTCGCAATAGACTTCAACAATTAAAAAAAATATCAATAGCAACAATTAG
AATTCTATTATAGAAACAACCTATTAGCAAGATAAATTCATAAATGGTGCAATAATTGCA
ATTAGGTCTCAAAATCTAGCTGTTAAGTATATTGTTGACCATATTAAGGAAATAACAGAC
AGAAATTTGTCTGAAGCTTATGTAAGAAAGATATGGGATCATTGGCTTAAATACAAAGAT
ATTTACTTCTCTTCAACACATAAAACAAGACTCTACACAAAGCATGAAGCCATTCAACTT
TATGATGCTATTTTTAAGCAATATAATCCAACTAACTAATCTTAAAGTATTATCCCTAAA
ATCAAAGAAGTATTGTAAAAACATGCTATAACAAAAAAATTGTTTATCAACTACTCACAA
TTCGAAGAAGTCAAAGGACTAGATACTGAAGAAACATATCTCTCAATTAAAGAAGCTATA
GACTCAGGTTAGTAATCTGATGAGCCAAAATGGATTGATGTCAGTGTAAGAATGATGATT
TCAGCAAGCTAGTTTGAATACAAGAAACAGATTGAAATTCAATGTTTTAATAAACTTAAA
AATTATTTGCAATTTTCTTGTATATAATTGGTTGGTGTAGTTATGGGTGAAACACTTAAT
GGAAGAGGAAAATTCATTTATATCCATTGTAGATATGATCAGTTAATACTAAAGAGGAAC
AAGTACTTGAAAAGATTCTTGCTTGAAGTTATTCCTGACTGGTTTTATTCTTATAACTGT
GTGCATAGTCAAAAAATTGCTATTTTTAAGTACTCTAAATTTTTATATCTCTAACGCAAG
ATACTTGAGGAGTAAGAAAGAAAAAGATTTATTCCTAATCCAGTCAATCCTCAGATAAAT
CAAACCAATCGTTTTAACAATTCTAGTGCTATAAATACCTCCCTTAACATTTCCTATCAA
TCATCTGCTGTTCCTAATACGAGTAGTATACCTCTGAACAGTTTATTCTGA>TTHERM_00675850(gene)
TTAATTAAAGGATACAAAAACAAACTGAAATAATTATATTATTATTAATTTATTCTAAAT
AAAAATTAATTTACCCTTTAAAAAAAGAAAAATAATTAATTATAAAAGACCCAACCCTTT
TTAATACAAAGAGCAGCGAGTATCTTCGTTTGAAATTTCTTTTAATACCTCCTATTTTAT
ACTCTATTTGATACCTCCTTAGATAGTCAATCCATTTATTAGGTTTTTGCCATACATTTG
ATACCTCCTCCTTAAATACTATTTCTCAATCTTAAATAATTCCAAGCTGTAAATAGATCA
TTGAATCGATCCCACTCTACTTAAATTCCTAAATATTTAAGACTCAAATGAAATCTCATT
CAAAGAAATACCAAGTAACAAATAATTTAGATGCTGACAATGAATTATATCCTCAATCAC
AAATTTAGAGTTCCAATAATGATTAGTAAACTCAACTGAACAGTCAAGAAGAAAATAACC
CTGATATGATTGAAACTGAAGATATAAACTTTAATCCAAATAACTCCAATAAACCAACAA
ATAGTTCTTATAAAATTCAAACAATAAACTTAGAGAATAACAATCTAGGTGAAAATACTT
CGCAATAGACTTCAACAATTAAAAAAAATATCAATAGCAACAATTAGAATTCTATTATAG
AAACAACCTATTAGCAAGATAAATTCATAAATGGTGCAATAATTGCAATTAGGTCTCAAA
ATCTAGCTGTTAAGTATATTGTTGACCATATTAAGGAAATAACAGACAGAAATTTGTCTG
AAGCTTATGTAAGAAAGATATGGGATCATTGGCTTAAATACAAAGATATTTACTTCTCTT
CAACACATAAAACAAGACTCTACACAAAGCATGAAGCCATTCAACTTTATGATGCTATTT
TTAAGCAATATAATCCAACTAACTAATCTTAAAGTATTATCCCTAAAATCAAAGAAGTAT
TGTAAAAACATGCTATAACAAAAAAATTGTTTATCAACTACTCACAATTCGAAGAAGTCA
AAGGACTAGATACTGAAGAAACATATCTCTCAATTAAAGAAGCTATAGACTCAGGTTAGT
AATCTGATGAGCCAAAATGGATTGATGTCAGTGTAAGAATGATGATTTCAGCAAGCTAGT
TTGAATACAAGAAACAGATTGAAATTCAATGTTTTAATAAACTTAAAAATTATTTGCAAT
TTTCTTGTATATAATTGGTTGGTGTAGTTATGGGTGAAACACTTAATGGAAGAGGAAAAT
TCATTTATATCCATTGTAGATATGATCAGTTAATACTAAAGAGGAACAAGTACTTGAAAA
GATTCTTGCTTGAAGTTATTCCTGACTGGTTTTATTCTTATAACTGTGTGCATAGTCAAA
AAATTGCTATTTTTAAGTACTCTAAATTTTTATATCTCTAACGCAAGATACTTGAGGAGT
AAGAAAGAAAAAGATTTATTCCTAATCCAGTCAATCCTCAGATAAATCAAACCAATCGTT
TTAACAATTCTAGTGCTATAAATACCTCCCTTAACATTTCCTATCAATCATCTGCTGTTC
CTAATACGAGTAGTATACCTCTGAACAGTTTATTCTGATAGAACAATATTAAATTTAAAT
ATATAAAGAAAATAAATATGTATCCTATTAAATTTTTTGTTTCATTTGATTTAATT>TTHERM_00675850(protein)
MKSHSKKYQVTNNLDADNELYPQSQIQSSNNDQQTQLNSQEENNPDMIETEDINFNPNNS
NKPTNSSYKIQTINLENNNLGENTSQQTSTIKKNINSNNQNSIIETTYQQDKFINGAIIA
IRSQNLAVKYIVDHIKEITDRNLSEAYVRKIWDHWLKYKDIYFSSTHKTRLYTKHEAIQL
YDAIFKQYNPTNQSQSIIPKIKEVLQKHAITKKLFINYSQFEEVKGLDTEETYLSIKEAI
DSGQQSDEPKWIDVSVRMMISASQFEYKKQIEIQCFNKLKNYLQFSCIQLVGVVMGETLN
GRGKFIYIHCRYDQLILKRNKYLKRFLLEVIPDWFYSYNCVHSQKIAIFKYSKFLYLQRK
ILEEQERKRFIPNPVNPQINQTNRFNNSSAINTSLNISYQSSAVPNTSSIPLNSLF