Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00016170Standard Name
HHT3 (Histone H Three)Aliases
PreTt11924 | 1.m00972 | g25486Description
HHT3 histone H3.4; Histone minor H3 variant, H3.3; replication independent replacement variant found primarily in macronuclei; constitutively expressed; not essential for cell growth; Histone H3/CENP-AGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- macronucleus (TAS) | GO:0031039
- micronucleus (IDA) | GO:0031040
- prospore membrane spindle pole body attachment site (IEA) | GO:0070057
Molecular Function
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- CAP-Gly domain binding (IEA) | GO:0071794
- obsolete bacterial-type RNA polymerase activity (IEA) | GO:0001061
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase (ATP) activity (IEA) | GO:0004612
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- phosphoglycerate dehydrogenase activity (IEA) | GO:0004617
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
Biological Process
- cellular response to dopamine (IEA) | GO:1903351
- donor selection (IEA) | GO:0007535
- fibroblast growth factor receptor signaling pathway involved in mammary gland specification (IEA) | GO:0060595
- germ-line processes downstream of sex determination signal (IEA) | GO:0007547
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- inactivation of recombination (HML) (IEA) | GO:0007537
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- lateral sprouting from an epithelium (IEA) | GO:0060601
- mammary gland specification (IEA) | GO:0060594
- mammary placode formation (IEA) | GO:0060596
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete extracellular carbohydrate transport (IEA) | GO:0006859
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- ommochrome biosynthetic process (IEA) | GO:0006727
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of dendrite extension (IEA) | GO:1903861
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of bile acid metabolic process (IEA) | GO:1904251
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of phosphorus utilization (IEA) | GO:0006795
- response to methamphetamine hydrochloride (IEA) | GO:1904313
- somatic processes downstream of sex determination signal (IEA) | GO:0007546
- striated muscle myosin thick filament assembly (IEA) | GO:0071688
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- transcription by RNA polymerase V (IEA) | GO:0001060
 Domains
Domains
- ( PF00125 ) Core histone H2A/H2B/H3/H4
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
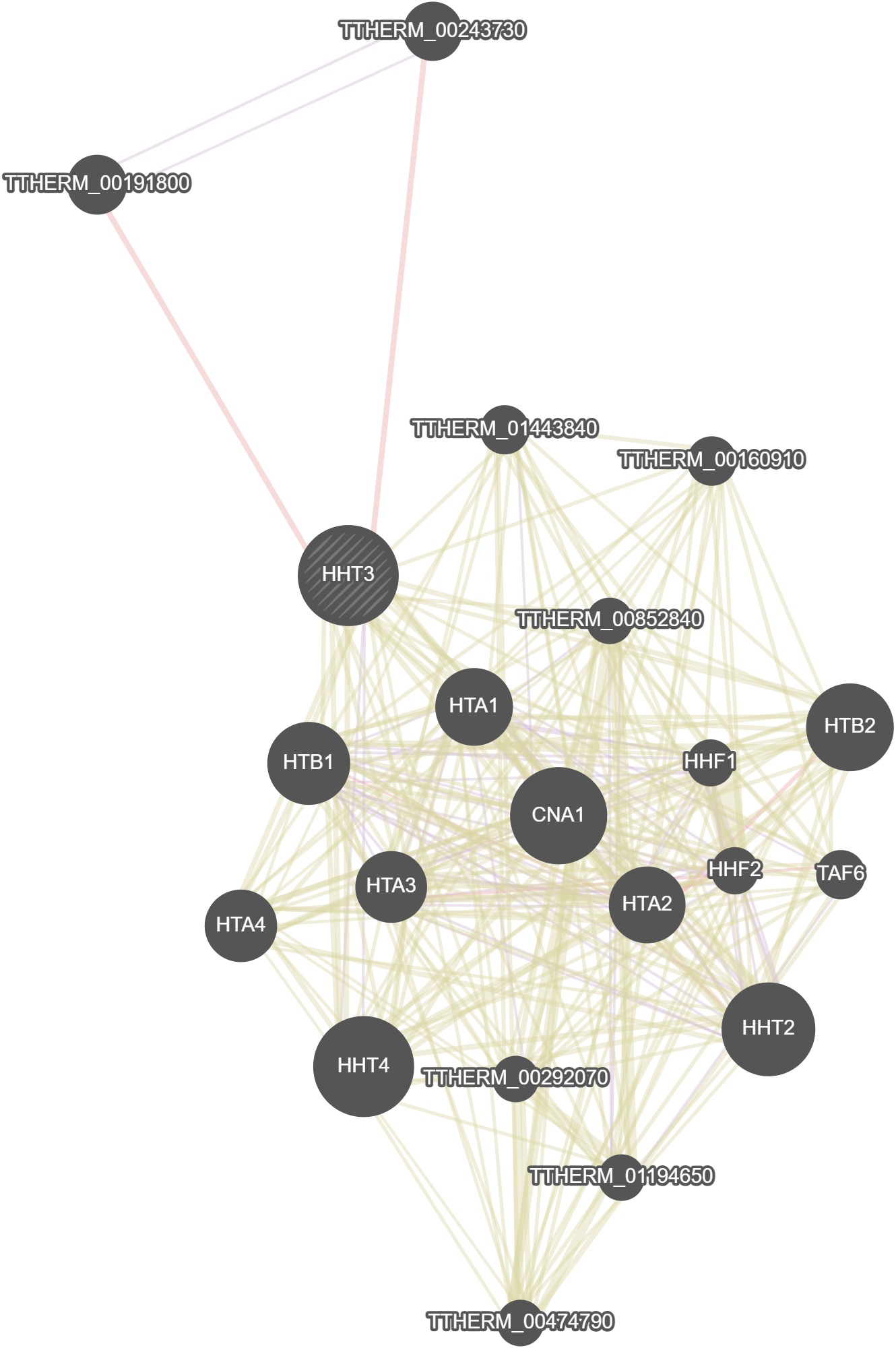
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
- ( SD01301 ) Macronucleus: Neo2 cassettes KO both HHT1 and HHT2
- ( SD01318 ) Macronucleus: KO of HHT3 with neo2 cassette
- ( SD01374 ) Macronucleus: N-terminal His- tag of HHT3, not sure if complete replacement
- ( SD01375 ) Macronucleus: N-terminal His- tag of HHT3
- ( SD01376 ) Macronucleus: N-terminal His- tag of HHT3
- ( SD01779 ) Macronucleus: C-terminal GFP- tag of HHT3
- ( SD01854 ) Micronucleus: GFP tagged HHT3 into HHT2 locus
- ( SD01855 ) Micronucleus: GFP tagged HHT3
- ( SD01856 ) Macronucleus: N-terminal His- tag of HHT3, not sure if complete replacement
- ( SD01857 ) Macronucleus: N-terminalFlag tagged HHT3 put into HHT4 locus, not sure if complete replacement
- ( SD01860 ) Micronucleus: Neo2 KO of HHT1 and HHT3
- ( SD01861 ) Micronucleus: Neo3 KO of HHT3
- ( SD01879 ) Micronucleus: Neo2 KO of HHT1 and HHT3
- ( SD01891 ) Macronucleus: N-terminal HA-His- tag of HHT3
- ( SD01892 ) Macronucleus: N-terminal HA tag of HHT3, not sure if complete replacement
- ( SD01896 ) Macronucleus: C-terminal GFP-tag of HHT3, not sure if complete replacement, into HHT2 locus
- ( SD01897 ) Macronucleus: C-terminal GFP- tag of HHT3, not sure if complete replacement
- ( SD01946 ) Micronucleus: GFP tagged HHT2 into HHT3 locus
- ( SD01947 ) Micronucleus: GFP tagged HHT2 into HHT3 locus
- ( SD01948 ) Micronucleus: GFP tagged HHT2 into HHT3 locus
- ( SD01950 ) Micronucleus: GFP tagged HHT3
- ( SD01951 ) Micronucleus: GFP tagged HHT3
- ( SD01952 ) Micronucleus: GFP tagged HHT3
- ( SD01953 ) Micronucleus: C-terminal GFP- tag of HHT3, into HHT2 locus
- ( SD01954 ) Micronucleus: C-terminal GFP- tag of HHT3, into HHT2 locus
- ( SD02213 ) Micronucleus: HHT3 KO with neo2, HHT4 KO with bsr1
- ( SD02214 ) Micronucleus: HHT3 KO with neo2, HHT4 KO with bsr1
- ( SD02215 ) Micronucleus: HHT3 KO with neo2, HHT4 KO with bsr1
- ( SD02302 ) Micronucleus: GFP tagged HHT2 into HHT3 locus
- ( SD02303 ) Micronucleus: C-terminal GFP- tag of HHT3, into HHT2 locus
- ( SD02314 ) Macronucleus: N-terminal His- tag of HHT3, not sure if complete replacement
- ( SD02556 ) Micronucleus: HHT3 KO with neo2, HHT4 KO with bsr1 Macronucleus: HHT3 KO with neo2, HHT4 KO with bsr1
- ( SD02557 ) Micronucleus: HHT3 KO with neo2, HHT4 KO with bsr1 Macronucleus: HHT3 KO with neo2, HHT4 KO with bsr1
- ( SD02643 ) Macronucleus: N-terminal HA-His- tag of HHT3
- ( SD02644 ) Macronucleus: N-terminal HA tag of HHT3
- ( SD02645 ) Macronucleus: N-terminal HA tag of HHT3
- ( SD02646 ) Macronucleus: N-terminal HA tag of HHT3
- ( SD02647 ) Macronucleus: N-terminal HA tag of HHT3
- ( SD02648 ) Macronucleus: N-terminal HA tag of HHT3
- ( SD02666 ) Macronucleus: neo 2 replaces HHT3
- ( SD02667 ) Micronucleus: HHT3 KO with neo2, HHT4 KO with bsr1 Macronucleus: HHT3 KO with neo2, HHT4 KO with bsr1
- ( SD02668 ) Micronucleus: HHT3 KO with neo2, HHT4 KO with bsr1 Macronucleus: HHT3 KO with neo2, HHT4 KO with bsr1
- ( SD02669 ) Macronucleus: Neo2 cassettes KO HHT3, partial KO of HHT2
- ( SD02670 ) Macronucleus: N-terminal His- tag of HHT3
- ( SD02720 ) Macronucleus: HHT3 with GFP c-terminal tag replaces the MTT1 coding region
- ( SD02742 ) Micronucleus: neo2 into HHT3 Macronucleus: neo2 into HHT3
- ( SD02743 ) Micronucleus: neo2 into HHT3 Macronucleus: neo2 into HHT3
- ( SD02793 ) Macronucleus: Neo2 in place of most of HHT3 coding region
- ( SD02794 ) Micronucleus: GFP into HHT3 Macronucleus: GFP into HHT3 with neo2 cassette
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:37024975: Nabeel-Shah S, Garg J, Ashraf K, Jeyapala R, Lee H, Petrova A, Burns JD, Pu S, Zhang Z, Greenblatt JF, Pearlman RE, Lambert JP, Fillingham J (2023) Multilevel interrogation of H3.3 reveals a primordial role in transcription regulation. Epigenetics & chromatin 16:10
- Ref:16908532: Cui B, Liu Y, Gorovsky MA (2006) Deposition and function of histone H3 variants in Tetrahymena thermophila. Molecular and cellular biology 26(20):7719-30
- Ref:15196465: Mochizuki K, Gorovsky MA (2004) Small RNAs in genome rearrangement in Tetrahymena. Current opinion in genetics & development 14(2):181-7
- Ref:14755052: Liu Y, Mochizuki K, Gorovsky MA (2004) Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proceedings of the National Academy of Sciences of the United States of America 101(6):1679-84
- Ref:14661947: Poux AN, Marmorstein R (2003) Molecular basis for Gcn5/PCAF histone acetyltransferase selectivity for histone and nonhistone substrates. Biochemistry 42(49):14366-74
- Ref:14536085: Clements A, Poux AN, Lo WS, Pillus L, Berger SL, Marmorstein R (2003) Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Molecular cell 12(2):461-73
- Ref:12665578: Ren Q, Gorovsky MA (2003) The nonessential H2A N-terminal tail can function as an essential charge patch on the H2A.Z variant N-terminal tail. Molecular and cellular biology 23(8):2778-89
- Ref:12522258: Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science (New York, N.Y.) 299(5607):716-9
- Ref:12455963: Duharcourt S, Yao MC (2002) Role of histone deacetylation in developmentally programmed DNA rearrangements in Tetrahymena thermophila. Eukaryotic cell 1(2):293-303
- Ref:12351775: Jenuwein T (2002) Molecular biology. An RNA-guided pathway for the epigenome. Science (New York, N.Y.) 297(5590):2215-8
- Ref:12123289: Waterborg JH (2002) Dynamics of histone acetylation in vivo. A function for acetylation turnover? Biochemistry and cell biology = Biochimie et biologie cellulaire 80(3):363-78
- Ref:12297044: Taverna SD, Coyne RS, Allis CD (2002) Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell 110(6):701-11
- Ref:11994311: Langer MR, Fry CJ, Peterson CL, Denu JM (2002) Modulating acetyl-CoA binding in the GCN5 family of histone acetyltransferases. The Journal of biological chemistry 277(30):27337-44
- Ref:11972045: Dou Y, Gorovsky MA (2002) Regulation of transcription by H1 phosphorylation in Tetrahymena is position independent and requires clustered sites. Proceedings of the National Academy of Sciences of the United States of America 99(9):6142-6
- Ref:11891286: Shang Y, Song X, Bowen J, Corstanje R, Gao Y, Gaertig J, Gorovsky MA (2002) A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proceedings of the National Academy of Sciences of the United States of America 99(6):3734-9
- Ref:11784863: Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P (2002) Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Molecular and cellular biology 22(3):874-85
- Ref:11751634: Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes & development 15(24):3286-95
- Ref:11467741: Green GR (2001) Phosphorylation of histone variant regions in chromatin: unlocking the linker? Biochemistry and cell biology = Biochimie et biologie cellulaire 79(3):275-87
- Ref:11135657: Tan S (2001) One HAT size fits all? Nature structural biology 8(1):8-10
- Ref:10975520: De Souza CP, Osmani AH, Wu LP, Spotts JL, Osmani SA (2000) Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell 102(3):293-302
- Ref:10975519: Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, Lin R, Smith MM, Allis CD (2000) Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102(3):279-91
- Ref:10503205: Wiley EA, Mizzen CA, Allis CD (2000) Isolation and characterization of in vivo modified histones and an activity gel assay for identification of histone acetyltransferases. Methods in cell biology 62( ):379-94
- Ref:11112280: Trievel RC, Li FY, Marmorstein R (2000) Application of a fluorescent histone acetyltransferase assay to probe the substrate specificity of the human p300/CBP-associated factor. Analytical biochemistry 287(2):319-28
- Ref:10199406: Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD (1999) Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97(1):99-109
- Ref:10549296: Dou Y, Mizzen CA, Abrams M, Allis CD, Gorovsky MA (1999) Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Molecular cell 4(4):641-7
- Ref:10611321: Strahl BD, Ohba R, Cook RG, Allis CD (1999) Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proceedings of the National Academy of Sciences of the United States of America 96(26):14967-72
- Ref:10551870: Karrer KM, VanNuland TA (1999) Nucleosome positioning is independent of histone H1 in vivo. The Journal of biological chemistry 274(46):33020-4
- Ref:10536141: Hettmann C, Soldati D (1999) Cloning and analysis of a Toxoplasma gondii histone acetyltransferase: a novel chromatin remodelling factor in Apicomplexan parasites. Nucleic acids research 27(22):4344-52
- Ref:9343391: Yu L, Gorovsky MA (1997) Constitutive expression, not a particular primary sequence, is the important feature of the H3 replacement variant hv2 in Tetrahymena thermophila. Molecular and cellular biology 17(11):6303-10
- Ref:8121802: Thatcher TH, MacGaffey J, Bowen J, Horowitz S, Shapiro DL, Gorovsky MA (1994) Independent evolutionary origin of histone H3.3-like variants of animals and Tetrahymena. Nucleic acids research 22(2):180-6
 Sequences
Sequences
>TTHERM_00016170(coding)
ATGGCTAGAACTAAATAAACTGCTAGAAAGTCAACTGGTGTTAAGGCTCCTAGAAAACAA
CTCGCTACCAAGGCTGCCAGAAAGTCTGCCCCCGTCTCTGGTGGTGTCAAGAAGCCCCAT
AAATTCAGACCTGGTACCGTCGCCTTGAGAGAAATCAGAAAGTATTAAAAGACTACTGAC
TTACTTATTAGAAAGCTCCCCTTCTAAAGACTTGTCAGAGATATTGCTATGGAAATGAAG
AGTGATATCAGATTCCAATCCCAAGCTATCCTTGCCCTCCAAGAAGCCGCTGAAGCCTAC
CTCGTCGGTTTATTCGAAGATACCAACCTCTGCGCTATCCACGCTAGAAGAGTTACTATT
ATGACCAAGGATCTCCACCTTGCTAGAAGAATTAGAGGAGAAAGATTCTGA>TTHERM_00016170(gene)
AAACAAAATTAAATTAAAAAAAAATTTAATCAAAAAAAAAAACCTCAAAGTTTTTTGAAA
AAAGCAATTAAAAAAATAATTTTCACACAAAACAGTAACAAAAATGGCTAGAACTAAATA
AACTGCTAGAAAGTCAACTGGTGTTAAGGCTCCTAGAAAACAACTCGCTACCAAGGCTGC
CAGAAAGTCTGCCCCCGTCTCTGGTGGTGTCAAGAAGCCCCATAAATTCAGACCTGGTAC
CGTCGCCTTGAGAGAAATCAGAAAGTATTAAAAGACTACTGACTTACTTATTAGAAAGCT
CCCCTTCTAAAGACTTGTCAGAGATATTGCTATGGAAATGAAGAGTGATATCAGATTCCA
ATCCCAAGCTATCCTTGCCCTCCAAGAAGCCGCTGAAGCCTACCTCGTCGGTTTATTCGA
AGATACCAACCTCTGCGCTATCCACGCTAGAAGAGTTACTATTATGACCAAGGATCTCCA
CCTTGCTAGAAGAATTAGAGGAGAAAGATTCTGAGAGCTCTCTCTAATAATTAACTTAAT
ATATAATACACACATATAATATAATGTTTTCTTATTATGGGAATGCAGCTTGCATCTTGA
TGCTTTTTACTCTCTTTGATTATAAATAAACTTCATTGATTCATACGGATACATAAAATC
ATACTCTATATCAAGTACTTTATTCACTCATTCATTGATGAGTGAGTGAATGAATGAAAA
AACATGGGTGGTTGATTAACTTATTAATTAATTTAATATAAAGACTGCATATATGCCAAT
CAACATGCATACATGCATGAAATGTAATTAATTGGTCCTGACAAATAAATTATGTATAAA
TAAATAAATAAAACTATTAAAACGCATTGAGCCAAAAACAGCTTGCATACTGCATCTTCC
AATTACTTGTTTCATACACTCTTATATAAATTTAAATTAGTGGTATCAAATCACATAAAA
TATATATAAATATATATTATGTATAATCTTAATTTTATTGCTAATCATCTCTTTCAATTG
AAAAAGCTTATCCAAAAA>TTHERM_00016170(protein)
MARTKQTARKSTGVKAPRKQLATKAARKSAPVSGGVKKPHKFRPGTVALREIRKYQKTTD
LLIRKLPFQRLVRDIAMEMKSDIRFQSQAILALQEAAEAYLVGLFEDTNLCAIHARRVTI
MTKDLHLARRIRGERF