Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00016200Standard Name
HHT4 (Histone H Three)Aliases
PreTt11927 | 1.m00975Description
HHT4 histone H3.4; histone H3.3, putative; Histone H3/CENP-AGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- prospore membrane spindle pole body attachment site (IEA) | GO:0070057
Molecular Function
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- CAP-Gly domain binding (IEA) | GO:0071794
- obsolete bacterial-type RNA polymerase activity (IEA) | GO:0001061
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase (ATP) activity (IEA) | GO:0004612
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- phosphoglycerate dehydrogenase activity (IEA) | GO:0004617
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
Biological Process
- cellular response to dopamine (IEA) | GO:1903351
- donor selection (IEA) | GO:0007535
- fibroblast growth factor receptor signaling pathway involved in mammary gland specification (IEA) | GO:0060595
- germ-line processes downstream of sex determination signal (IEA) | GO:0007547
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- inactivation of recombination (HML) (IEA) | GO:0007537
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- lateral sprouting from an epithelium (IEA) | GO:0060601
- mammary gland specification (IEA) | GO:0060594
- mammary placode formation (IEA) | GO:0060596
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete extracellular carbohydrate transport (IEA) | GO:0006859
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- ommochrome biosynthetic process (IEA) | GO:0006727
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of dendrite extension (IEA) | GO:1903861
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of bile acid metabolic process (IEA) | GO:1904251
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of phosphorus utilization (IEA) | GO:0006795
- response to methamphetamine hydrochloride (IEA) | GO:1904313
- somatic processes downstream of sex determination signal (IEA) | GO:0007546
- striated muscle myosin thick filament assembly (IEA) | GO:0071688
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- transcription by RNA polymerase V (IEA) | GO:0001060
 Domains
Domains
- ( PF00125 ) Core histone H2A/H2B/H3/H4
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
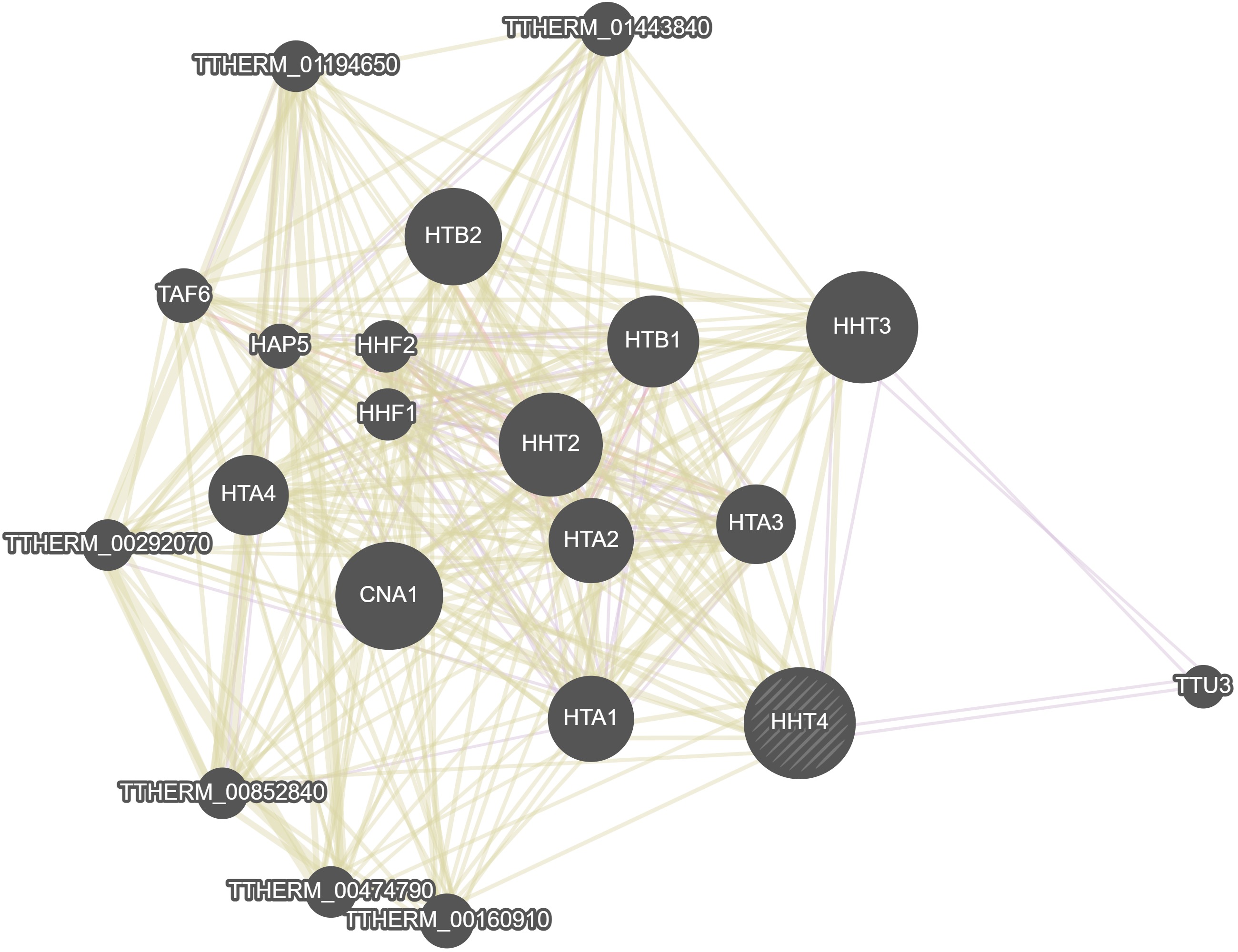
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:37024975: Nabeel-Shah S, Garg J, Ashraf K, Jeyapala R, Lee H, Petrova A, Burns JD, Pu S, Zhang Z, Greenblatt JF, Pearlman RE, Lambert JP, Fillingham J (2023) Multilevel interrogation of H3.3 reveals a primordial role in transcription regulation. Epigenetics & chromatin 16:10
- Ref:16908532: Cui B, Liu Y, Gorovsky MA (2006) Deposition and function of histone H3 variants in Tetrahymena thermophila. Molecular and cellular biology 26(20):7719-30
 Sequences
Sequences
>TTHERM_00016200(coding)
ATGGTGATTCCTTTATTATAAACACTCTTTTTTTTTGAAAATAAGTTTATAATATTTTCA
AAACAAAATAAAATTAAATTAAAAAAATTTTCATCAAAACAAAAAAATAATTTACACAAA
ACAGAAAAGAAAATGGCTAGAACTAAATAAACTGCTAGAAAGTCAACTAGTATTAAGGCT
CCTAGAAAACAATTGGCTGCCAAGGCTGCCAGAAAGTCTGCCCCCATCTCTGGTGGTATC
AAAAAGCCCCATAAATTCAGACCTGGTACCGTTGCCTTGAGAGAAATCAGAAAGTATTAA
AAGACTACTGACTTACTTATTAGAAAGCTCCCCTTCTAAAGACTTGTCAGAGATATTGCT
ATGGAAATGAAGAGCGATATCAGATTCCAATCACAAGCTATCCTTGCCCTCCAAGAAGCT
GCTGAAGCCTACCTAGTAGGTTTATTCGAAGATACCAACCTCTGCGCTATCCACGCTAGA
AGAGTTACTATTATGACCAAGGATCTCCACCTTGCTAGAAGAATTAGAGGAGAAAGATTC
TGA>TTHERM_00016200(gene)
CACTTTAAGCATATATTATTGATTAACTTACATAATTGGAATAGAAATGGTGATTCCTTT
ATTATAAACACTCTTTTTTTTTGAAAATAAGTTTATAATATTTTCAAAACAAAATAAAAT
TAAATTAAAAAAATTTTCATCAAAACAAAAAAATAATTTACACAAAACAGAAAAGAAAAT
GGCTAGAACTAAATAAACTGCTAGAAAGTCAACTAGTATTAAGGCTCCTAGAAAACAATT
GGCTGCCAAGGCTGCCAGAAAGTCTGCCCCCATCTCTGGTGGTATCAAAAAGCCCCATAA
ATTCAGACCTGGTACCGTTGCCTTGAGAGAAATCAGAAAGTATTAAAAGACTACTGACTT
ACTTATTAGAAAGCTCCCCTTCTAAAGACTTGTCAGAGATATTGCTATGGAAATGAAGAG
CGATATCAGATTCCAATCACAAGCTATCCTTGCCCTCCAAGAAGCTGCTGAAGCCTACCT
AGTAGGTTTATTCGAAGATACCAACCTCTGCGCTATCCACGCTAGAAGAGTTACTATTAT
GACCAAGGATCTCCACCTTGCTAGAAGAATTAGAGGAGAAAGATTCTGAGAGCTCTCTCT
AATAATTAACTTAATATATAATACACACATATAATATAATGTTTTCTTATTATGGGAATC
ATTTAGTTTGTTTTACATAATAACTAAATTATTTATTTAATATGAGGTTTTTAAAATAAA
TATTTATAAAATAAAATTTTTTTGAAAAAAACATTAGTTA>TTHERM_00016200(protein)
MVIPLLQTLFFFENKFIIFSKQNKIKLKKFSSKQKNNLHKTEKKMARTKQTARKSTSIKA
PRKQLAAKAARKSAPISGGIKKPHKFRPGTVALREIRKYQKTTDLLIRKLPFQRLVRDIA
MEMKSDIRFQSQAILALQEAAEAYLVGLFEDTNLCAIHARRVTIMTKDLHLARRIRGERF