Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00069680Standard Name
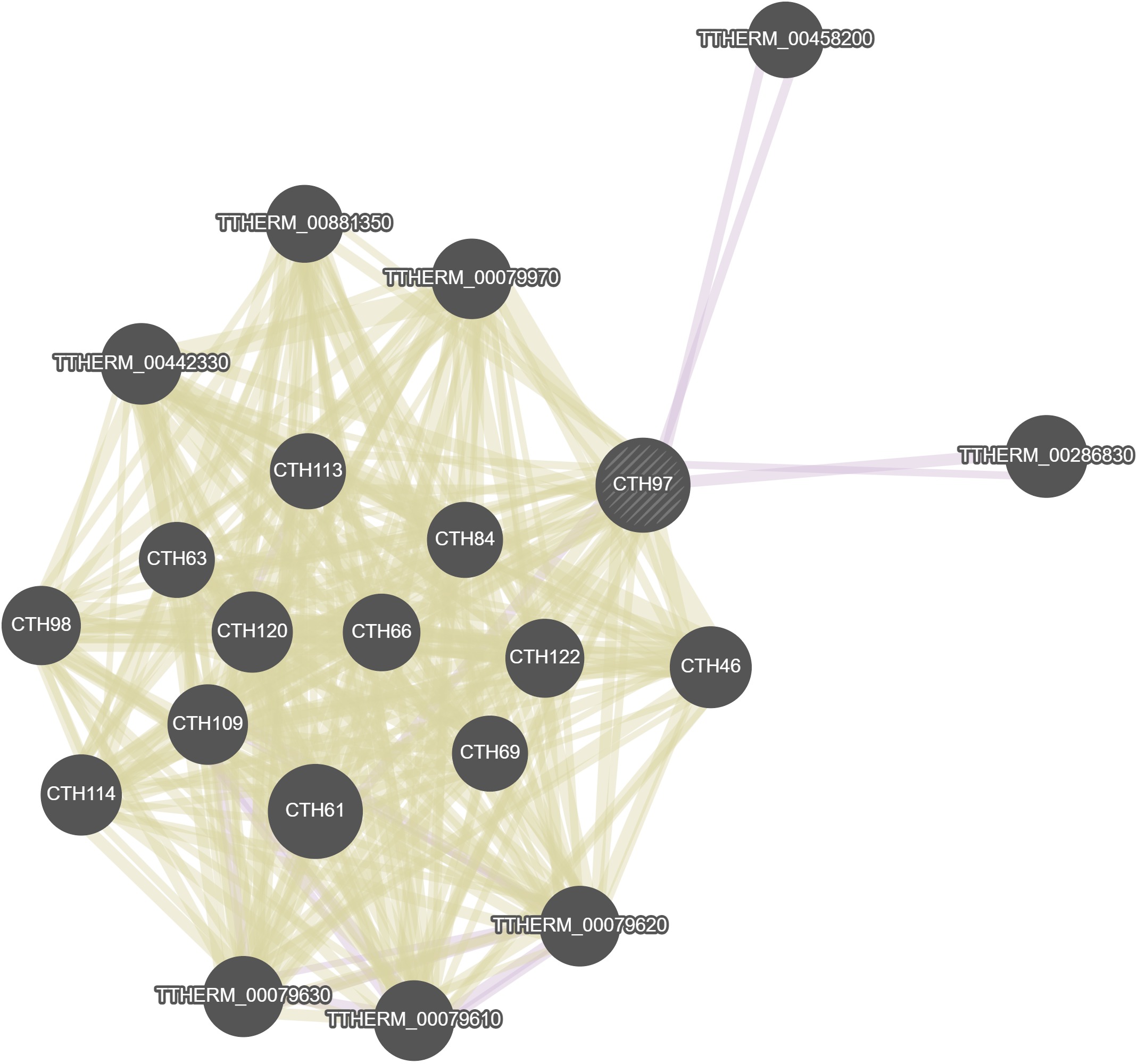
CTH97 (CaTHepsin 97)Aliases
PreTt13257 | 4.m00656 | 3707.m00292Description
CTH97 papain family cysteine protease; Peptidase C1AGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- Dbf4-dependent protein kinase complex (IEA) | GO:0031431
- obsolete integral component of mitochondrial membrane (IEA) | GO:0032592
- THO complex (IEA) | GO:0000347
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- Ac-Asp-Glu binding (IEA) | GO:1904492
- cytoskeletal anchor activity (IEA) | GO:0008093
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- ferric triacetylfusarinine C:proton symporter activity (IEA) | GO:0015346
- folic acid transmembrane transporter activity (IEA) | GO:0008517
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- insulin-like growth factor II binding (IEA) | GO:0031995
- interleukin-10 receptor binding (IEA) | GO:0005141
- kappa-type opioid receptor binding (IEA) | GO:0031851
- N-acetyl-beta-D-glucosaminide beta-(1,3)-galactosyltransferase activity (IEA) | GO:0008499
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete glycine-, glutamate-, thienylcyclohexylpiperidine binding (IEA) | GO:0008500
- obsolete peroxisome-assembly ATPase activity (IEA) | GO:0008573
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- organic anion transmembrane transporter activity (IEA) | GO:0008514
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- small ribosomal subunit rRNA binding (IEA) | GO:0070181
- sodium-independent organic anion transmembrane transporter activity (IEA) | GO:0015347
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- sucrose transmembrane transporter activity (IEA) | GO:0008515
- threonine-type peptidase activity (IEA) | GO:0070003
- thyroid hormone transmembrane transporter activity (IEA) | GO:0015349
- tumor necrosis factor receptor binding (IEA) | GO:0005164
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
Biological Process
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- auditory receptor cell fate commitment (IEA) | GO:0009912
- baroreceptor detection of decreased arterial stretch (IEA) | GO:0003024
- bronchiole development (IEA) | GO:0060435
- bronchus development (IEA) | GO:0060433
- CD24 biosynthetic process (IEA) | GO:0035724
- CD8-positive, alpha-beta cytotoxic T cell differentiation (IEA) | GO:0002308
- cell migration in diencephalon (IEA) | GO:0061381
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular component organization or biogenesis (IEA) | GO:0071840
- cellular response to dopamine (IEA) | GO:1903351
- DNA damage response (IEA) | GO:0006974
- DNA recombinase mediator complex assembly (IEA) | GO:1903871
- embryonic brain development (IEA) | GO:1990403
- epithelium development (IEA) | GO:0060429
- epithelium regeneration (IEA) | GO:1990399
- eukaryotic translation initiation factor 2B complex assembly (IEA) | GO:1905173
- extraembryonic membrane development (IEA) | GO:1903867
- glycolytic process from glycerol (IEA) | GO:0061613
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- humoral immune response (IEA) | GO:0006959
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- inferior colliculus development (IEA) | GO:0061379
- interferon-kappa secretion (IEA) | GO:0072650
- intraciliary retrograde transport (IEA) | GO:0035721
- lung basal cell differentiation (IEA) | GO:0060508
- lung connective tissue development (IEA) | GO:0060427
- macrophage derived foam cell differentiation (IEA) | GO:0010742
- modulation by organism of defense-related calcium ion flux in other organism involved in symbiotic interaction (IEA) | GO:0052301
- modulation by organism of defense-related nitric oxide production in other organism involved in symbiotic interaction (IEA) | GO:0052302
- muscle contraction (IEA) | GO:0006936
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of antimicrobial peptide production (IEA) | GO:0002785
- negative regulation of central B cell deletion (IEA) | GO:0002899
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of interleukin-4-mediated signaling pathway (IEA) | GO:1902215
- negative regulation of leukocyte cell-cell adhesion (IEA) | GO:1903038
- negative regulation of maintenance of meiotic sister chromatid cohesion, centromeric (IEA) | GO:2000710
- negative regulation of maintenance of mitotic sister chromatid cohesion, centromeric (IEA) | GO:2000719
- negative regulation of peripheral B cell deletion (IEA) | GO:0002909
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phospholipid translocation (IEA) | GO:0061093
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete induction by virus of host cell-cell fusion (IEA) | GO:0006948
- obsolete melanotic tumor response (IEA) | GO:0006969
- obsolete multi-organism cellular localization (IEA) | GO:1902581
- obsolete negative regulation of telomeric RNA transcription from RNA pol II promoter (IEA) | GO:1901581
- obsolete pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction (IEA) | GO:0052308
- obsolete positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response (IEA) | GO:0006990
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- obsolete sodium/potassium transport (IEA) | GO:0006815
- ommochrome biosynthetic process (IEA) | GO:0006727
- P granule disassembly (IEA) | GO:1903864
- palisade mesophyll development (IEA) | GO:1903866
- phagocytosis (IEA) | GO:0006909
- phenanthrene catabolic process via trans-9(R),10(R)-dihydrodiolphenanthrene (IEA) | GO:0018956
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation by organism of phytoalexin production in other organism involved in symbiotic interaction (IEA) | GO:0052329
- positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction (IEA) | GO:0052330
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of antimicrobial peptide biosynthetic process (IEA) | GO:0002807
- positive regulation of antimicrobial peptide secretion (IEA) | GO:0002796
- positive regulation of biosynthetic process of antibacterial peptides active against Gram-negative bacteria (IEA) | GO:0006964
- positive regulation of cap-independent translational initiation (IEA) | GO:1903679
- positive regulation of cell cycle phase transition (IEA) | GO:1901989
- positive regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation (IEA) | GO:2000704
- positive regulation of flower development (IEA) | GO:0009911
- positive regulation of macropinocytosis (IEA) | GO:1905303
- positive regulation of myofibroblast differentiation (IEA) | GO:1904762
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of peroxisome proliferator activated receptor signaling pathway (IEA) | GO:0035360
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of skeletal muscle satellite cell differentiation (IEA) | GO:1902726
- positive regulation of stress response to copper ion (IEA) | GO:1903855
- positive regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903691
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- prostate induction (IEA) | GO:0060514
- protein localization to old growing cell tip (IEA) | GO:1903858
- pteridine biosynthetic process (IEA) | GO:0006728
- pyrimidine deoxyribonucleotide biosynthetic process (IEA) | GO:0009221
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of antibacterial peptide biosynthetic process (IEA) | GO:0002808
- regulation of antisense RNA transcription (IEA) | GO:0060194
- regulation of biosynthetic process of antibacterial peptides active against Gram-positive bacteria (IEA) | GO:0002816
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of dense core granule biogenesis (IEA) | GO:2000705
- regulation of double fertilization forming a zygote and endosperm (IEA) | GO:0080155
- regulation of interferon-gamma secretion (IEA) | GO:1902713
- regulation of macrophage derived foam cell differentiation (IEA) | GO:0010743
- regulation of maintenance of mitotic sister chromatid cohesion, centromeric (IEA) | GO:2000718
- regulation of mitophagy (IEA) | GO:1901524
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of sprouting angiogenesis (IEA) | GO:1903670
- regulation of systemic arterial blood pressure by baroreceptor feedback (IEA) | GO:0003025
- regulation of systemic arterial blood pressure by carotid body chemoreceptor signaling (IEA) | GO:0003027
- regulation of turning behavior involved in mating (IEA) | GO:0061094
- regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903689
- response to osmotic stress (IEA) | GO:0006970
- response to stress (IEA) | GO:0006950
- single-organism intracellular transport (IEA) | GO:1902582
- smooth muscle contraction (IEA) | GO:0006939
- sperm aster formation (IEA) | GO:0035044
- sterol regulatory element binding protein cleavage involved in ER-nuclear sterol response pathway (IEA) | GO:0006992
- synaptic vesicle docking (IEA) | GO:0016081
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- vacuolar acidification (IEA) | GO:0007035
- vitamin A metabolic process (IEA) | GO:0006776
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00069680(coding)
ATGAAGTAGCATAACCAAGAAAAAAATAATAGTTATTAGATAGGTATGAATTAATTTTCG
GACCTTACAATAGAAGAATTTCAGTCTATTTCATTAAATCAATAACTATTTAATAGTGAA
TCAAGAAAATTAGAAAATATAAAAAATGAAAATTAATAAGCTGATTTTTACTTGCAACTT
CTCAAAACTAATGCAAGTAGCTTACCATAATAATTTGATTGGAGAAATTTAGGAAAAGTT
ACTCAAGTTAAAAATTAAGGTAACTGTGGTTCTTGTTGGGCATTTACTATAACTGGTCTA
TTTGAAAGCATAAATTTAATTAGAAATAAAACAGTTGAACTATATTCTGAATAAGAACTA
TTAGACTGTTCTTCAAACGGCATTTATAGAAATTCAGGTTGTTAAGGAGGTTGGCCACAT
TTAGCATTTGAATACTCCAAAAAAAATGGAATATCTCTTTCTTCATAATATCCATACAAA
GGTATATAAGAAAACTGCACAGTTAATTAGCAAACTAAAAAGGCTTTTTATCCTAGTTAG
CCTATTTAAATTTAAGCTGATTAAGAAAGTAACAAAATACAAATTATCAAATAACTACTT
TTAAATTCACCTCTAGCTGTTATAGTAGATGCCTCTAATTGGTCTAACTATAAATCAGGA
GTCTTTTCAAATTGCACTACATAATAAAATCATGTAGCTTTGCTAGTCGGATACACTAAC
GAAGGAAATTGGATAATTAAAAACTCTTGGGGGTCTGCTTGGGGTGAATCTGGGTACATA
ACCTTAAGCGCAAATAATGCCTGTGGTTTAGAAAATAATATTCTTGGATCTACAATAATT
TGA>TTHERM_00069680(gene)
CTAATTTCAGGCTATAAATTTTTCTATTAAACATAAAGCAAATGAAGTAGCATAACCAAG
AAAAAAATAATAGTTATTAGATAGGTATGAATTAATTTTCGGACCTTACAATAGAAGAAT
TTCAGTCTATTTCATTAAATCAATAACTATTTAATAGTGAATCAAGAAAATTAGAAAATA
TAAAAAATGAAAATTAATAAGCTGATTTTTACTTGCAACTTCTCAAAACTAATGCAAGTA
GCTTACCATAATAATTTGATTGGAGAAATTTAGGAAAAGTTACTCAAGTTAAAAATTAAG
GTAACTGTGGTTCTTGTTGGGCATTTACTATAACTGGTCTATTTGAAAGCATAAATTTAA
TTAGAAATAAAACAGTTGAACTATATTCTGAATAAGAACTATTAGACTGTTCTTCAAACG
GCATTTATAGAAATTCAGGTTGTTAAGGAGGTTGGCCACATTTAGCATTTGAATACTCCA
AAAAAAATGGAATATCTCTTTCTTCATAATATCCATACAAAGGTATATAAGAAAACTGCA
CAGTTAATTAGCAAACTAAAAAGGCTTTTTATCCTAGTTAGCCTATTTAAATTTAAGCTG
ATTAAGAAAGTAACAAAATACAAATTATCAAATAACTACTTTTAAATTCACCTCTAGCTG
TTATAGTAGATGCCTCTAATTGGTCTAACTATAAATCAGGAGTCTTTTCAAATTGCACTA
CATAATAAAATCATGTAGCTTTGCTAGTCGGATACACTAACGAAGGAAATTGGATAATTA
AAAACTCTTGGGGGTCTGCTTGGGGTGAATCTGGGTACATAACCTTAAGCGCAAATAATG
CCTGTGGTTTAGAAAATAATATTCTTGGATCTACAATAATTTGAAATGTTTTATTAACAA
ACAAAAGTACGTATAGTAAGAAAGTATATTAATTGATTTTAAAGCTGAAT>TTHERM_00069680(protein)
MKQHNQEKNNSYQIGMNQFSDLTIEEFQSISLNQQLFNSESRKLENIKNENQQADFYLQL
LKTNASSLPQQFDWRNLGKVTQVKNQGNCGSCWAFTITGLFESINLIRNKTVELYSEQEL
LDCSSNGIYRNSGCQGGWPHLAFEYSKKNGISLSSQYPYKGIQENCTVNQQTKKAFYPSQ
PIQIQADQESNKIQIIKQLLLNSPLAVIVDASNWSNYKSGVFSNCTTQQNHVALLVGYTN
EGNWIIKNSWGSAWGESGYITLSANNACGLENNILGSTII