Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00083670Standard Name
RAB65 (RAB GTPase 65)Aliases
PreTt16314 | 6.m00388 | 3836.m01984Description
RAB65 Ras family small GTPase; Novel Rab family protein; small monomeric GTPase that regulates membrane fusion and fission events; Small GTPase superfamily, RabGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- chloroplast ribosome (IEA) | GO:0043253
- extracellular membraneless organelle (IEA) | GO:0043264
- female pronucleus (IEA) | GO:0001939
- glutathione hydrolase complex (IEA) | GO:0061672
- laminin-8 complex (IEA) | GO:0043257
- MICOS complex (IEA) | GO:0061617
- obsolete ectoplasm (IEA) | GO:0043265
- obsolete integral component of mitochondrial membrane (IEA) | GO:0032592
- ubiquitin ligase complex (IEA) | GO:0000151
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-octaprenyl-6-methoxyphenol hydroxylase activity (IEA) | GO:0008681
- 3-hydroxybutyrate dehydrogenase activity (IEA) | GO:0003858
- alpha-humulene synthase activity (IEA) | GO:0080017
- Atg12 ligase activity (IEA) | GO:0061660
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- FAT10 ligase activity (IEA) | GO:0061661
- folate:monoatomic anion antiporter activity (IEA) | GO:0008518
- folic acid transmembrane transporter activity (IEA) | GO:0008517
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- insulin-like growth factor II binding (IEA) | GO:0031995
- kappa-type opioid receptor binding (IEA) | GO:0031851
- L-ascorbate:sodium symporter activity (IEA) | GO:0008520
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- mercury ion transmembrane transporter activity (IEA) | GO:0015097
- N-acetyl-beta-D-glucosaminide beta-(1,3)-galactosyltransferase activity (IEA) | GO:0008499
- obsolete 3-HSA hydroxylase activity (IEA) | GO:0034819
- obsolete 4-chloronitrobenzene nitroreductase activity (IEA) | GO:0052602
- obsolete aminoacetone:oxygen oxidoreductase(deaminating) activity (IEA) | GO:0052594
- obsolete delta-tocopherol cyclase activity (IEA) | GO:0052604
- obsolete endogenous peptide receptor activity (IEA) | GO:0008529
- obsolete NAD(P)+ transhydrogenase activity (IEA) | GO:0008746
- obsolete RNA polymerase I transcription regulator recruiting activity (IEA) | GO:0001186
- obsolete serpin (IEA) | GO:0004868
- obsolete sodium-translocating F-type ATPase activity (IEA) | GO:0016468
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylinositol-3-phosphate binding (IEA) | GO:0032266
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- Pup ligase activity (IEA) | GO:0061664
- replication fork barrier binding (IEA) | GO:0031634
- sodium:iodide symporter activity (IEA) | GO:0008507
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- sucrose:proton symporter activity (IEA) | GO:0008506
- threonine-type peptidase activity (IEA) | GO:0070003
- thrombospondin receptor activity (IEA) | GO:0070053
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
- UFM1 ligase activity (IEA) | GO:0061666
Biological Process
- bipolar cellular bud site selection (IEA) | GO:0007121
- budding cell apical bud growth (IEA) | GO:0007118
- budding cell bud growth (IEA) | GO:0007117
- budding cell isotropic bud growth (IEA) | GO:0007119
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to carbendazim (IEA) | GO:0072762
- cellular response to dsRNA (IEA) | GO:0071359
- cellular response to hydroxyurea (IEA) | GO:0072711
- chaperone-mediated protein folding (IEA) | GO:0061077
- chemokine (C-C motif) ligand 5 signaling pathway (IEA) | GO:0035689
- collagen metabolic process (IEA) | GO:0032963
- commissural neuron axon guidance (IEA) | GO:0071679
- epithelium regeneration (IEA) | GO:1990399
- glycolytic process from fructose through fructose-6-phosphate (IEA) | GO:0061616
- glycolytic process from glycerol (IEA) | GO:0061613
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- humoral immune response (IEA) | GO:0006959
- IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor (IEA) | GO:0002416
- immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor (IEA) | GO:0002415
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- interleukin-19 production (IEA) | GO:0032622
- interleukin-2 production (IEA) | GO:0032623
- L-ornithine transmembrane transport (IEA) | GO:1903352
- left horn of sinus venosus development (IEA) | GO:0061079
- lung morphogenesis (IEA) | GO:0060425
- macrophage derived foam cell differentiation (IEA) | GO:0010742
- mannosyl diphosphorylinositol ceramide metabolic process (IEA) | GO:0006676
- mitotic cytokinetic cell separation (IEA) | GO:1902409
- mitotic nuclear division (IEA) | GO:0007067
- modulation by organism of defense-related cell wall thickening in other organism involved in symbiotic interaction (IEA) | GO:0052300
- negative regulation of chemokine-mediated signaling pathway (IEA) | GO:0070100
- negative regulation of epithelial cell proliferation involved in lung morphogenesis (IEA) | GO:2000795
- negative regulation of glycine secretion, neurotransmission (IEA) | GO:1904625
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phospholipid translocation (IEA) | GO:0061093
- negative regulation of phytoalexin metabolic process (IEA) | GO:0052321
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process (IEA) | GO:0062026
- neutral lipid metabolic process (IEA) | GO:0006638
- nucleocytoplasmic transport (IEA) | GO:0006913
- obsolete activation of MAPK activity (IEA) | GO:0000187
- obsolete dynamin family protein polymerization involved in mitochondrial fission (IEA) | GO:0003374
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete multi-organism cellular localization (IEA) | GO:1902581
- obsolete negative regulation of cGMP catabolic process (IEA) | GO:0030830
- obsolete negative regulation of histone H3-K27 methylation (IEA) | GO:0061086
- obsolete phagosome formation (IEA) | GO:0006912
- obsolete regulation of histone H3-K79 methylation (IEA) | GO:2001160
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete reproduction (IEA) | GO:0000003
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- ommochrome biosynthetic process (IEA) | GO:0006727
- P granule assembly (IEA) | GO:1903863
- phagocytosis (IEA) | GO:0006909
- phagocytosis, engulfment (IEA) | GO:0006911
- phenanthrene catabolic process via trans-9(R),10(R)-dihydrodiolphenanthrene (IEA) | GO:0018956
- photosynthetic NADP+ reduction (IEA) | GO:0009780
- plant-type cell wall modification (IEA) | GO:0009827
- polyamine acetylation (IEA) | GO:0032917
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation (IEA) | GO:2000704
- positive regulation of forebrain neuron differentiation (IEA) | GO:2000979
- positive regulation of prostaglandin secretion involved in immune response (IEA) | GO:0061078
- positive regulation of venous endothelial cell fate commitment (IEA) | GO:2000789
- pre-replicative complex assembly involved in cell cycle DNA replication (IEA) | GO:1902299
- pronephric glomerulus morphogenesis (IEA) | GO:0035775
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- prostate induction (IEA) | GO:0060514
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of antisense RNA transcription (IEA) | GO:0060194
- regulation of chitin metabolic process (IEA) | GO:0032882
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of lens fiber cell differentiation (IEA) | GO:1902746
- regulation of microglial cell migration (IEA) | GO:1904139
- regulation of nucleus organization (IEA) | GO:1903353
- regulation of PERK-mediated unfolded protein response (IEA) | GO:1903897
- regulation of protein deneddylation (IEA) | GO:0060625
- regulation of protein localization to cell division site (IEA) | GO:1901900
- regulation of sprouting angiogenesis (IEA) | GO:1903670
- regulation of ubiquitin homeostasis (IEA) | GO:0010993
- single-organism intracellular transport (IEA) | GO:1902582
- striated muscle contraction (IEA) | GO:0006941
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
 Domains
Domains
- ( PF00071 ) Ras family
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
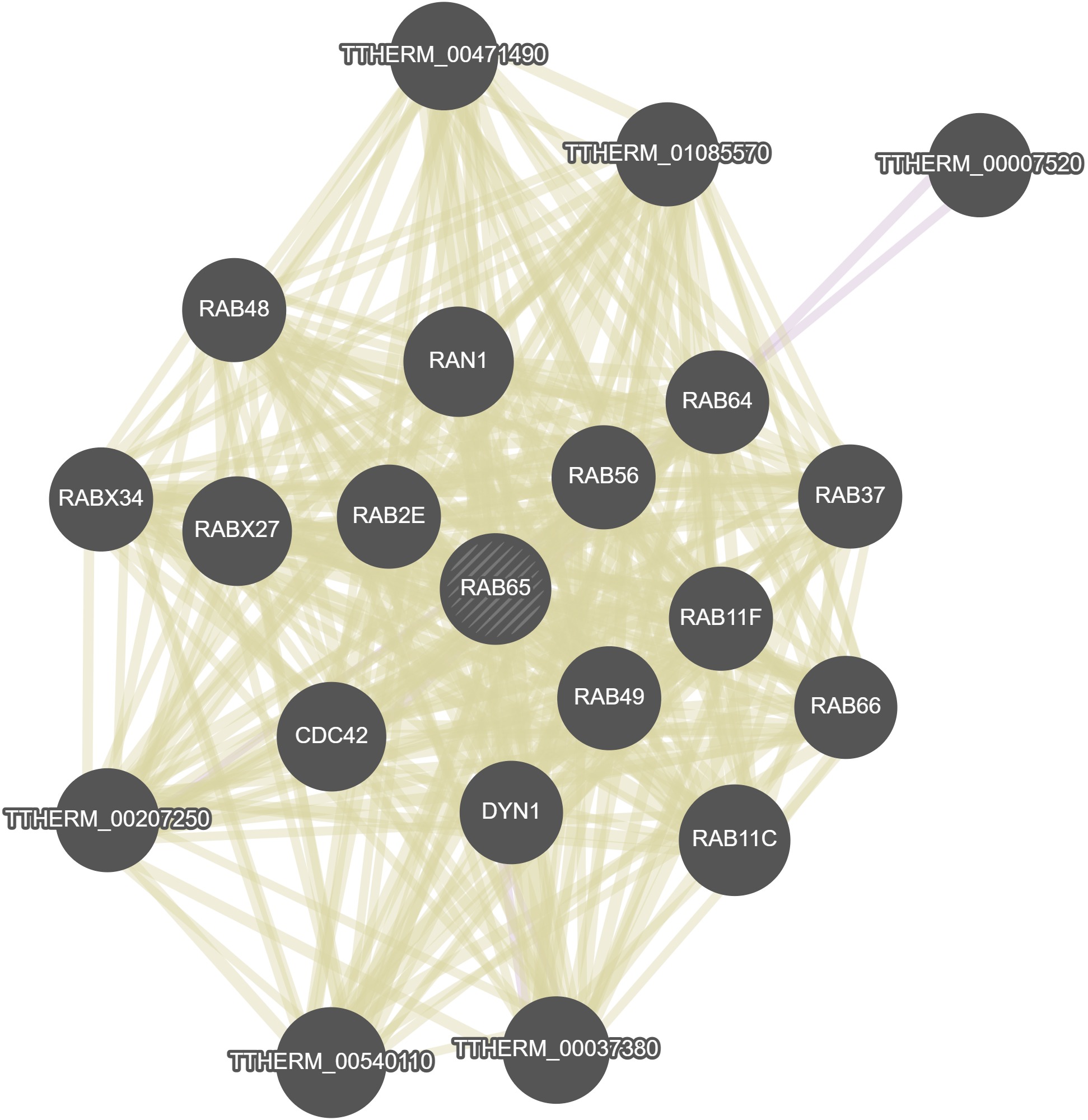
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:16933976: Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA,
 Sequences
Sequences
>TTHERM_00083670(coding)
ATGAGTTAGAATAAGGATGATTAGCAAATTTATTTTAAAATATTAATGATAGGCAACTCA
GCAGTTGGAAATACATGTCTATTGGTGAGATATTGCAATAGTGAATTTCCAAATAGTAGC
ATAATCACATTGGGTATAGATTTAAAAATAAAAAAACTTGAAATTAATAAAAAAAAGCTT
AATTTGCATATTTTGGATACAGCTAGTTAGGATAAATTTAGAACCATTACTCCAAATTAT
TTCAAAGCAGCTCATGGTATTTTAGTAGCCTATTCAGTAGAAGACTAAGAATCGTTTGAG
AAAGTAGGAAGCTGGATTTAATGCATAAAAGAAAATTTCGATAATGCTAATATATCTATA
TTATTAGTTGGAAATAAATCTGATTCAAAACAAAGAGTAGTTTCATTAGAATAGGGTGAA
AAGCTTGCTGAGTAATATAATATTCCTTTTTAGGAAACTAGTGCTCTTAATAATATTAAT
GTGAGCGAATCTTTTTATTAACTTGCTAAAATGTGCTTAGATTAGTATGAGATAAAAAGT
TCTACTAGGTCTCCTTAATCTATTGTACCAAAGAAAAAATAAGAAAATGGAGAAACTAAA
AAAAATTGTTGTTGA>TTHERM_00083670(gene)
TTGAAAAGAAAGTAAAGTAAAAGGTTATTTGTGAAAAATTGATTTATGAGTTAGAATAAG
GATGATTAGCAAATTTATTTTAAAATATTAATGATAGGCAACTCAGCAGTTGGAAATACA
TGTCTATTGGTGAGATATTGCAATAGTGAATTTCCAAATAGTAGCATAATCACATTGGGT
ATAGATTTAAAAATAAAAAAACTTGAAATTAATAAAAAAAAGCTTAATTTGCATATTTTG
GATACAGCTAGTTAGGATAAATTTAGAACCATTACTCCAAATTATTTCAAAGCAGCTCAT
GGTATTTTAGTAGCCTATTCAGTAGAAGACTAAGAATCGTTTGAGAAAGTAGGAAGCTGG
ATTTAATGCATAAAAGAAAATTTCGATAATGCTAATATATCTATATTATTAGTTGGAAAT
AAATCTGATTCAAAACAAAGAGTAGTTTCATTAGAATAGGGTGAAAAGCTTGCTGAGTAA
TATAATATTCCTTTTTAGGAAACTAGTGCTCTTAATAATATTAATGTGAGCGAATCTTTT
TATTAACTTGCTAAAATGTGCTTAGATTAGTATGAGATAAAAAGTTCTACTAGGTCTCCT
TAATCTATTGTACCAAAGAAAAAATAAGAAAATGGAGAAACTAAAAAAAATTGTTGTTGA
AAAGTTTAGCTTTTTTAAACTAAAATTTTCACGCCTATCCAACTATTCATCCATCAATCT
ATCTTCTATATTTAACAAAGCAGATAAATAAGTCTAAAAAACAATTTAATATAAAATATT
GCTAATTACTACAAATTTAAATGAATAATTAATTTCATTTATTCTAAATAATATTATATT
ATCATTAAAAAACAGTGTAATTAACTATTAATCAATTTTATTGTTATTTAAATTCATTTT
TTAATGTTTATATTCATTTTAAAATACTTCATTATCAATAAACCCAATAAAC>TTHERM_00083670(protein)
MSQNKDDQQIYFKILMIGNSAVGNTCLLVRYCNSEFPNSSIITLGIDLKIKKLEINKKKL
NLHILDTASQDKFRTITPNYFKAAHGILVAYSVEDQESFEKVGSWIQCIKENFDNANISI
LLVGNKSDSKQRVVSLEQGEKLAEQYNIPFQETSALNNINVSESFYQLAKMCLDQYEIKS
STRSPQSIVPKKKQENGETKKNCC