Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00112480Standard Name
THD15 (Tetrahymena Histone Deacetylase 15)Aliases
PreTt27062 | 8.m00322 | 3812.m02359Description
THD15 SIR2 family transcriptional regulator; Sir2-like histone deacetylase; suppresses DNA transcription by deacetylation of histones; NAD-dependent sirtuin protein deacylasesGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- alpha6-beta4 integrin-Shc-Grb2 complex (IEA) | GO:0070333
- CURI complex (IEA) | GO:0032545
- Dbf4-dependent protein kinase complex (IEA) | GO:0031431
- EGFR-Grb2-Sos complex (IEA) | GO:0070620
- gamma-catenin-TCF7L2 complex (IEA) | GO:0071665
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- mononeme (IEA) | GO:0070074
- obsolete chromosome segregation-directing complex (IEA) | GO:0043913
- obsolete integral component of mitochondrial membrane (IEA) | GO:0032592
- Shc-Grb2-Sos complex (IEA) | GO:0070619
- subrhabdomeral cisterna (IEA) | GO:0016029
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-keto-3-deoxy-L-rhamnonate aldolase activity (IEA) | GO:0106099
- 4-coumarate-CoA ligase activity (IEA) | GO:0016207
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- ABC-type amino acid transporter activity (IEA) | GO:0015424
- ABC-type oligosaccharide transporter activity (IEA) | GO:0015422
- acylglycerone-phosphate reductase (NADP+) activity (IEA) | GO:0000140
- alpha-humulene synthase activity (IEA) | GO:0080017
- antheraxanthin epoxidase activity (IEA) | GO:0052663
- apelin receptor activity (IEA) | GO:0060182
- arachidonate omega-hydroxylase activity (IEA) | GO:0052869
- arsenate reductase (glutaredoxin) activity (IEA) | GO:0008794
- axon guidance receptor activity (IEA) | GO:0008046
- cAMP-dependent protein kinase regulator activity (IEA) | GO:0008603
- CAP-Gly domain binding (IEA) | GO:0071794
- carboxymethylenebutenolidase activity (IEA) | GO:0008806
- carboxyvinyl-carboxyphosphonate phosphorylmutase activity (IEA) | GO:0008807
- cardiolipin synthase activity (IEA) | GO:0008808
- carnitine:acyl carnitine antiporter activity (IEA) | GO:0005476
- catechol O-methyltransferase activity (IEA) | GO:0016206
- cellulase activity (IEA) | GO:0008810
- Cys-tRNA(Pro) hydrolase activity (IEA) | GO:0043907
- cytoskeletal anchor activity (IEA) | GO:0008093
- dAMP binding (IEA) | GO:0032562
- endopeptidase regulator activity (IEA) | GO:0061135
- fatty acyl-[ACP] hydrolase activity (IEA) | GO:0016297
- ferric enterobactin:proton symporter activity (IEA) | GO:0015345
- ferric triacetylfusarinine C:proton symporter activity (IEA) | GO:0015346
- flap-structured DNA binding (IEA) | GO:0070336
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- glycogen synthase activity, transferring glucose-1-phosphate (IEA) | GO:0061547
- histone acetyltransferase binding (IEA) | GO:0035035
- inositol-1,4,5,6-tetrakisphosphate 2-kinase activity (IEA) | GO:0032942
- interferon receptor activity (IEA) | GO:0004904
- juvenile hormone epoxide hydrolase activity (IEA) | GO:0008096
- L-aspartate oxidase activity (IEA) | GO:0008734
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- lanosterol synthase activity (IEA) | GO:0000250
- lipopolysaccharide heptosyltransferase activity (IEA) | GO:0008920
- lipopolysaccharide N-acetylglucosaminyltransferase activity (IEA) | GO:0008917
- low voltage-gated potassium channel auxiliary protein activity (IEA) | GO:0015458
- metarhodopsin binding (IEA) | GO:0016030
- methane monooxygenase [NAD(P)H] activity (IEA) | GO:0015049
- microtubule severing ATPase activity (IEA) | GO:0008568
- mitochondrial single subunit type RNA polymerase activity (IEA) | GO:0001065
- neoxanthin synthase activity (IEA) | GO:0034020
- obsolete aspartic-type signal peptidase activity (IEA) | GO:0009049
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete D-lysine oxidase activity (IEA) | GO:0043912
- obsolete DNA topoisomerase IV activity (IEA) | GO:0008724
- obsolete gp130 (IEA) | GO:0004898
- obsolete myristoyl-[acyl-carrier-protein] hydrolase activity (IEA) | GO:0016295
- obsolete phosphatidylinositol 3-kinase, class I, catalyst activity (IEA) | GO:0015072
- obsolete proprotein convertase 2 activator activity (IEA) | GO:0016484
- obsolete prostaglandin/thromboxane transporter activity (IEA) | GO:0015348
- obsolete red-sensitive opsin (IEA) | GO:0015061
- obsolete sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity (IEA) | GO:0052591
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete type II protein secretor activity (IEA) | GO:0015447
- obsolete X-opioid receptor activity (IEA) | GO:0015051
- obsolete zinc, iron permease activity (IEA) | GO:0015342
- phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity (IEA) | GO:0016314
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase (ATP) activity (IEA) | GO:0004612
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- phosphoglycerate dehydrogenase activity (IEA) | GO:0004617
- pyrimidine nucleotide-sugar transmembrane transporter activity (IEA) | GO:0015165
- quercitrin binding (IEA) | GO:2001227
- R-linalool synthase activity (IEA) | GO:0034008
- rhamnulose-1-phosphate aldolase activity (IEA) | GO:0008994
- RNA polymerase III activity (IEA) | GO:0001056
- Ser(Gly)-tRNA(Ala) hydrolase activity (IEA) | GO:0043908
- siderophore uptake transmembrane transporter activity (IEA) | GO:0015344
- siderophore-iron transmembrane transporter activity (IEA) | GO:0015343
- single-subunit type RNA polymerase binding (IEA) | GO:0001050
- sodium-independent organic anion transmembrane transporter activity (IEA) | GO:0015347
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- Swi5-Sfr1 complex binding (IEA) | GO:1905334
- thyroid hormone transmembrane transporter activity (IEA) | GO:0015349
- transcription regulatory region nucleic acid binding (IEA) | GO:0001067
- type 2 neuropeptide Y receptor binding (IEA) | GO:0031843
- type 3 melanocortin receptor binding (IEA) | GO:0031781
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
- tyramine receptor activity (IEA) | GO:0008226
Biological Process
- 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902634
- 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process (IEA) | GO:1902633
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- absorption of visible light (IEA) | GO:0016038
- activation of Ral GTPase activity (IEA) | GO:0032859
- adrenomedullin receptor signaling pathway (IEA) | GO:1990410
- apoptotic process (IEA) | GO:0006915
- aromatic amino acid metabolic process (IEA) | GO:0009072
- aspartate family amino acid biosynthetic process (IEA) | GO:0009067
- autophagosome organization (IEA) | GO:1905037
- bronchiole development (IEA) | GO:0060435
- bronchus development (IEA) | GO:0060433
- bronchus morphogenesis (IEA) | GO:0060434
- bud dilation involved in lung branching (IEA) | GO:0060503
- canalicular bile acid transport (IEA) | GO:0015722
- carbon utilization (IEA) | GO:0015976
- carboxylic acid transmembrane transport (IEA) | GO:1905039
- carotenoid metabolic process (IEA) | GO:0016116
- cell growth involved in cardiac muscle cell development (IEA) | GO:0061049
- cell migration in diencephalon (IEA) | GO:0061381
- cell proliferation involved in embryonic placenta development (IEA) | GO:0060722
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to biotin starvation (IEA) | GO:1990383
- cellular response to dopamine (IEA) | GO:1903351
- cellular response to fluoride (IEA) | GO:1902618
- cellular response to homocysteine (IEA) | GO:1905375
- cellular response to vanadate(3-) (IEA) | GO:1902439
- cerebral cortex development (IEA) | GO:0021987
- chaperone-mediated protein folding (IEA) | GO:0061077
- chloroplast fission (IEA) | GO:0010020
- chondrocyte hypertrophy (IEA) | GO:0003415
- coenzyme A biosynthetic process (IEA) | GO:0015937
- cytotoxic T cell pyroptotic cell death (IEA) | GO:1902483
- detection of stimulus involved in Dma1-dependent checkpoint (IEA) | GO:1902419
- diadenosine tetraphosphate catabolic process (IEA) | GO:0015967
- dorsal motor nucleus of vagus nerve structural organization (IEA) | GO:0035763
- endochondral bone growth (IEA) | GO:0003416
- endoderm development (IEA) | GO:0007492
- energy derivation by oxidation of reduced inorganic compounds (IEA) | GO:0015975
- epithelium development (IEA) | GO:0060429
- eugenol catabolic process (IEA) | GO:0042856
- eukaryotic translation initiation factor 2B complex assembly (IEA) | GO:1905173
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- gamma-tubulin complex assembly (IEA) | GO:1902481
- general transcription from RNA polymerase II promoter (IEA) | GO:0032568
- glomerular capillary formation (IEA) | GO:0072104
- glycolytic process from glycerol (IEA) | GO:0061613
- hair cycle process (IEA) | GO:0022405
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- imaginal disc eversion (IEA) | GO:0007561
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- inactivation of recombination (HML) (IEA) | GO:0007537
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- inferior colliculus development (IEA) | GO:0061379
- interleukin-12 production (IEA) | GO:0032615
- interleukin-13 production (IEA) | GO:0032616
- interleukin-28A secretion (IEA) | GO:0072628
- intra-Golgi vesicle-mediated transport (IEA) | GO:0006891
- intraciliary retrograde transport (IEA) | GO:0035721
- juxtaglomerular apparatus development (IEA) | GO:0072051
- L-alanine catabolic process (IEA) | GO:0042853
- lamellipodium morphogenesis (IEA) | GO:0072673
- laminaritriose transport (IEA) | GO:2001097
- lateral ganglionic eminence cell proliferation (IEA) | GO:0022018
- light absorption (IEA) | GO:0016037
- lipid transport across blood-brain barrier (IEA) | GO:1990379
- lung connective tissue development (IEA) | GO:0060427
- lung epithelium development (IEA) | GO:0060428
- lung growth (IEA) | GO:0060437
- lung pattern specification process (IEA) | GO:0060432
- lung saccule development (IEA) | GO:0060430
- luteal cell differentiation (IEA) | GO:1903728
- macrophage derived foam cell differentiation (IEA) | GO:0010742
- maltotetraose transport (IEA) | GO:2001099
- mammary gland branching involved in pregnancy (IEA) | GO:0060745
- mesonephric juxtaglomerular apparatus development (IEA) | GO:0061212
- mesonephros development (IEA) | GO:0001823
- mitochondrial fusion (IEA) | GO:0008053
- mitochondrial membrane organization (IEA) | GO:0007006
- mitotic cell cycle checkpoint signaling (IEA) | GO:0007093
- mitotic cytokinetic cell separation (IEA) | GO:1902409
- mitotic nuclear division (IEA) | GO:0007067
- mitotic spindle elongation (IEA) | GO:0000022
- myelination in peripheral nervous system (IEA) | GO:0022011
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of axo-dendritic protein transport (IEA) | GO:1905127
- negative regulation of axon guidance (IEA) | GO:1902668
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of chemokine (C-C motif) ligand 20 production (IEA) | GO:1903885
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of dendritic cell dendrite assembly (IEA) | GO:2000548
- negative regulation of dipeptide transmembrane transport (IEA) | GO:2001149
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of epithelial tube formation (IEA) | GO:1905277
- negative regulation of ferulate catabolic process (IEA) | GO:1901467
- negative regulation of flavonoid biosynthetic process (IEA) | GO:0009964
- negative regulation of hindgut contraction (IEA) | GO:0060451
- negative regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070108
- negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator (IEA) | GO:1902239
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of late endosome to lysosome transport (IEA) | GO:1902823
- negative regulation of myeloid dendritic cell chemotaxis (IEA) | GO:2000528
- negative regulation of neutrophil migration (IEA) | GO:1902623
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phytoalexin metabolic process (IEA) | GO:0052321
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of protein localization to presynapse (IEA) | GO:1905385
- negative regulation of protein maturation (IEA) | GO:1903318
- negative regulation of response to gamma radiation (IEA) | GO:2001229
- negative regulation of response to wounding (IEA) | GO:1903035
- negative regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process (IEA) | GO:0062026
- negative regulation of thiamine biosynthetic process (IEA) | GO:0070624
- negative regulation of ubiquinone biosynthetic process (IEA) | GO:1904774
- negative regulation of vasculogenesis (IEA) | GO:2001213
- negative regulation of wound healing (IEA) | GO:0061045
- neutral lipid metabolic process (IEA) | GO:0006638
- nitrate transmembrane transport (IEA) | GO:0015706
- nuclear DNA replication termination (IEA) | GO:1902317
- obsolete 2-methylthio-N-6-(cis-hydroxy)isopentenyl adenosine-tRNA biosynthesis (IEA) | GO:0002195
- obsolete activation of MAPK activity (IEA) | GO:0000187
- obsolete activation of MAPKK during sporulation (sensu Saccharomyces) (IEA) | GO:0000204
- obsolete activation of pro-apoptotic gene products (IEA) | GO:0008633
- obsolete actomyosin contractile ring disassembly (IEA) | GO:1902621
- obsolete age dependent accumulation of genetic damage (IEA) | GO:0007570
- obsolete aging (IEA) | GO:0007568
- obsolete appressorium septum formation (IEA) | GO:0075033
- obsolete canonical Wnt signaling pathway involved in mesonephric nephron development (IEA) | GO:0061293
- obsolete extracellular carbohydrate transport (IEA) | GO:0006859
- obsolete fertilization, exchange of chromosomal proteins (IEA) | GO:0035042
- obsolete maintenance of dosage compensation (IEA) | GO:0007551
- obsolete modification by symbiont of host morphology or physiology via protein secreted by Sec complex (IEA) | GO:0085024
- obsolete modification by symbiont of host morphology or physiology via protein secreted by Tat complex (IEA) | GO:0085025
- obsolete modulation of peptidase activity in other organism involved in symbiotic interaction (IEA) | GO:0052198
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete multi-organism cellular localization (IEA) | GO:1902581
- obsolete multi-organism intracellular transport (IEA) | GO:1902583
- obsolete negative regulation of acetylcholine-gated cation channel activity (IEA) | GO:1903049
- obsolete negative regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity (IEA) | GO:1904218
- obsolete negative regulation of cytochrome-c oxidase activity (IEA) | GO:1905376
- obsolete negative regulation of histone H3-K27 methylation (IEA) | GO:0061086
- obsolete negative regulation of recombination within rDNA repeats (IEA) | GO:0000020
- obsolete negative regulation of RNA binding transcription factor activity (IEA) | GO:1905256
- obsolete negative regulation of telomeric RNA transcription from RNA pol II promoter (IEA) | GO:1901581
- obsolete nuclear division involved in appressorium formation (IEA) | GO:0075034
- obsolete phagosome formation (IEA) | GO:0006912
- obsolete positive chemotaxis in environment of other organism involved in symbiotic interaction (IEA) | GO:0052221
- obsolete positive regulation by host of symbiont defense response (IEA) | GO:0052197
- obsolete positive regulation of adhesion of symbiont to host epithelial cell (IEA) | GO:1905228
- obsolete positive regulation of nucleic acid-templated transcription (IEA) | GO:1903508
- obsolete positive regulation of telomeric RNA transcription from RNA pol II promoter (IEA) | GO:1901582
- obsolete positive regulation of transcription from RNA polymerase II promoter in response to nitrosative stress (IEA) | GO:0061403
- obsolete positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response (IEA) | GO:0006990
- obsolete process resulting in tolerance to hydrolysate (IEA) | GO:1990368
- obsolete regulation of 1-deoxy-D-xylulose-5-phosphate synthase activity (IEA) | GO:1902395
- obsolete regulation of apoptosis by sphingosine-1-phosphate signaling pathway (IEA) | GO:0003377
- obsolete regulation of DNA N-glycosylase activity (IEA) | GO:1902544
- obsolete regulation of dynamin family protein polymerization involved in membrane fission (IEA) | GO:0003375
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- obsolete sodium/potassium transport (IEA) | GO:0006815
- obsolete synthesis of RNA primer involved in premeiotic DNA replication (IEA) | GO:1902980
- ocellus pigment granule organization (IEA) | GO:0008058
- olfactory lobe development (IEA) | GO:0021988
- ommochrome biosynthetic process (IEA) | GO:0006727
- organelle organization (IEA) | GO:0006996
- otic placode development (IEA) | GO:1905040
- P granule assembly (IEA) | GO:1903863
- P granule disassembly (IEA) | GO:1903864
- penetration peg formation (IEA) | GO:0075053
- pericyte cell migration (IEA) | GO:1905351
- peripheral tolerance induction (IEA) | GO:0002465
- peripheral tolerance induction to self antigen (IEA) | GO:0002466
- phagocytosis (IEA) | GO:0006909
- phagocytosis, engulfment (IEA) | GO:0006911
- photoreceptor cell morphogenesis (IEA) | GO:0008594
- phyllotactic patterning (IEA) | GO:0060771
- plant-type cell wall modification (IEA) | GO:0009827
- polarity specification of anterior/posterior axis (IEA) | GO:0009949
- poly(3-hydroxyalkanoate) biosynthetic process (IEA) | GO:0042621
- positive regulation by organism of phytoalexin production in other organism involved in symbiotic interaction (IEA) | GO:0052329
- positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction (IEA) | GO:0052330
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of antifungal innate immune response (IEA) | GO:1905036
- positive regulation of branching involved in lung morphogenesis (IEA) | GO:0061047
- positive regulation of cap-independent translational initiation (IEA) | GO:1903679
- positive regulation of cardiac muscle contraction (IEA) | GO:0060452
- positive regulation of cell adhesion involved in sprouting angiogenesis (IEA) | GO:0106090
- positive regulation of chaperone-mediated protein folding (IEA) | GO:1903646
- positive regulation of cutin biosynthetic process (IEA) | GO:1901959
- positive regulation of deacetylase activity (IEA) | GO:0090045
- positive regulation of defense response to bacterium, incompatible interaction (IEA) | GO:1902479
- positive regulation of dendrite extension (IEA) | GO:1903861
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of epidermal growth factor receptor signaling pathway involved in heart process (IEA) | GO:1905284
- positive regulation of epithelial cell proliferation involved in lung bud dilation (IEA) | GO:0060504
- positive regulation of forebrain neuron differentiation (IEA) | GO:2000979
- positive regulation of glycoprotein biosynthetic process involved in immunological synapse formation (IEA) | GO:2000526
- positive regulation of I-kappaB phosphorylation (IEA) | GO:1903721
- positive regulation of inner ear receptor cell differentiation (IEA) | GO:2000982
- positive regulation of intracellular lipid transport (IEA) | GO:0032379
- positive regulation of macropinocytosis (IEA) | GO:1905303
- positive regulation of membrane hyperpolarization (IEA) | GO:1902632
- positive regulation of mesonephros development (IEA) | GO:0061213
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of peroxisome proliferator activated receptor signaling pathway (IEA) | GO:0035360
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of prostaglandin secretion involved in immune response (IEA) | GO:0061078
- positive regulation of protein maturation (IEA) | GO:1903319
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process (IEA) | GO:0062027
- positive regulation of sterol transport (IEA) | GO:0032373
- positive regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903691
- potassium ion transmembrane transport (IEA) | GO:0071805
- primary lung bud formation (IEA) | GO:0060431
- priming of cellular response to stress (IEA) | GO:0080136
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- proteasome storage granule assembly (IEA) | GO:1902906
- protein autosumoylation (IEA) | GO:1990466
- protein localization to cell division site involved in cytokinesis, actomyosin contractile ring assembly (IEA) | GO:1902575
- protein localization to mitotic spindle (IEA) | GO:1902480
- protein localization to old growing cell tip (IEA) | GO:1903858
- protein processing (IEA) | GO:0016485
- prothoracic disc morphogenesis (IEA) | GO:0007470
- proximal/distal pattern formation involved in nephron development (IEA) | GO:0072047
- pyrimidine deoxyribonucleotide biosynthetic process (IEA) | GO:0009221
- pyrimidine nucleotide salvage (IEA) | GO:0032262
- pyrimidine ribonucleotide catabolic process (IEA) | GO:0009222
- quercetin catabolic process (IEA) | GO:1901733
- quercetin metabolic process (IEA) | GO:1901732
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of antisense RNA transcription (IEA) | GO:0060194
- regulation of asexual reproduction (IEA) | GO:1903664
- regulation of axo-dendritic protein transport (IEA) | GO:1905126
- regulation of branching involved in lung morphogenesis (IEA) | GO:0061046
- regulation of cap-independent translational initiation (IEA) | GO:1903677
- regulation of cell growth involved in cardiac muscle cell development (IEA) | GO:0061050
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of chemokine (C-C motif) ligand 20 production (IEA) | GO:1903884
- regulation of cholesterol transport (IEA) | GO:0032374
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of dipeptide transmembrane transport (IEA) | GO:2001148
- regulation of ecdysteroid secretion (IEA) | GO:0007555
- regulation of gastric motility (IEA) | GO:1905333
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of hematopoietic progenitor cell differentiation (IEA) | GO:1901532
- regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070107
- regulation of L-arginine import into cell (IEA) | GO:1902826
- regulation of late endosome to lysosome transport (IEA) | GO:1902822
- regulation of leukocyte cell-cell adhesion (IEA) | GO:1903037
- regulation of long-chain fatty acid import across plasma membrane (IEA) | GO:0010746
- regulation of macrophage derived foam cell differentiation (IEA) | GO:0010743
- regulation of membrane lipid metabolic process (IEA) | GO:1905038
- regulation of opsin-mediated signaling pathway (IEA) | GO:0022400
- regulation of PERK-mediated unfolded protein response (IEA) | GO:1903897
- regulation of phosphatidylcholine biosynthetic process (IEA) | GO:2001245
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of plant epidermal cell differentiation (IEA) | GO:1903888
- regulation of protein localization to cell division site involved in cell separation after cytokinesis (IEA) | GO:1905391
- regulation of protein modification by small protein conjugation or removal (IEA) | GO:1903320
- regulation of raffinose metabolic process (IEA) | GO:0080091
- regulation of RNA binding transcription factor activity (IEA) | GO:1905255
- regulation of siRNA-mediated facultative heterochromatin formation (IEA) | GO:1902802
- regulation of sprouting angiogenesis (IEA) | GO:1903670
- regulation of T cell costimulation (IEA) | GO:2000523
- regulation of TOR signaling (IEA) | GO:0032006
- regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903689
- regulatory T cell apoptotic process (IEA) | GO:1902482
- release of sequestered calcium ion into cytosol by endoplasmic reticulum (IEA) | GO:1903514
- response to D-galactose (IEA) | GO:1905377
- response to mycotoxin (IEA) | GO:0010046
- response to sterol depletion (IEA) | GO:0006991
- retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum (IEA) | GO:0006890
- sesquiterpenoid biosynthetic process (IEA) | GO:0016106
- single-organism intracellular transport (IEA) | GO:1902582
- sperm aster formation (IEA) | GO:0035044
- sperm DNA decondensation (IEA) | GO:0035041
- spermatogonial cell division (IEA) | GO:0007284
- spindle assembly involved in male meiosis I (IEA) | GO:0007054
- sterol metabolic process (IEA) | GO:0016125
- sterol regulatory element binding protein cleavage involved in ER-nuclear sterol response pathway (IEA) | GO:0006992
- stilbene biosynthetic process (IEA) | GO:0009811
- substrate-dependent cell migration, cell attachment to substrate (IEA) | GO:0006931
- superior colliculus development (IEA) | GO:0061380
- synaptic vesicle docking (IEA) | GO:0016081
- synaptic vesicle priming (IEA) | GO:0016082
- T cell inhibitory signaling pathway (IEA) | GO:0002770
- T-helper 1 cell diapedesis (IEA) | GO:0035688
- terpenoid catabolic process (IEA) | GO:0016115
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- trachea submucosa development (IEA) | GO:0061152
- transcription by RNA polymerase V (IEA) | GO:0001060
- tricarboxylic acid transport (IEA) | GO:0006842
- triterpenoid catabolic process (IEA) | GO:0016105
- tRNA import into mitochondrion (IEA) | GO:0016031
- tyramine secretion (IEA) | GO:0061545
- vacuolar acidification (IEA) | GO:0007035
- vesicle coating (IEA) | GO:0006901
- xanthophyll biosynthetic process (IEA) | GO:0016123
- xanthophyll catabolic process (IEA) | GO:0016124
 Domains
Domains
- ( PF02146 ) Sir2 family
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
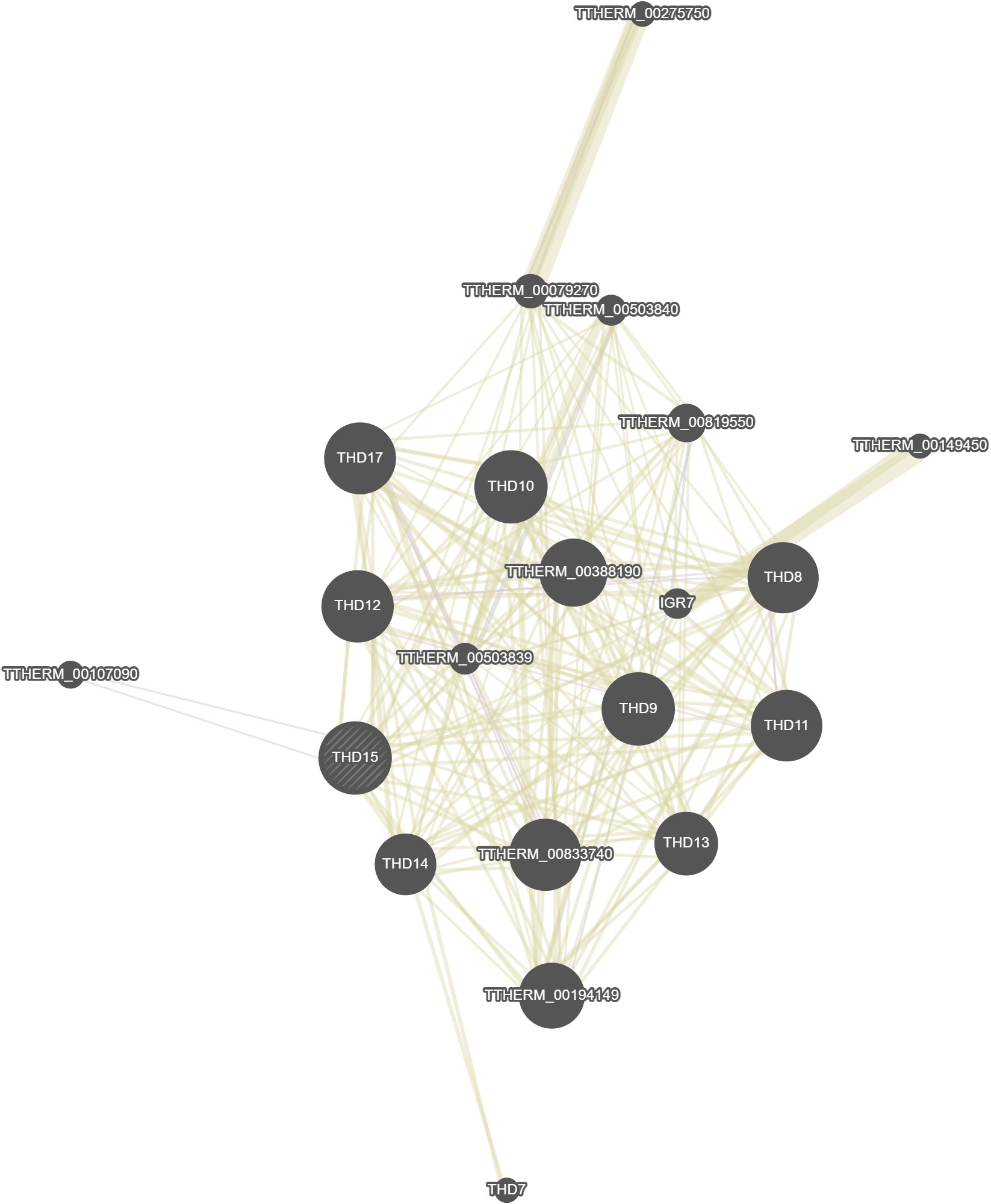
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:18178773: Smith JJ, Torigoe SE, Maxson J, Fish LC, Wiley EA (2008) A class II histone deacetylase acts on newly synthesized histones in Tetrahymena. Eukaryotic cell 7(3):471-82
 Sequences
Sequences
>TTHERM_00112480(coding)
ATGCATTTAAAAACACTCTTCAAATCATTAATTACTCCAAGTAAGTAAATTACTTTGAGA
ATATCTAACGTATAGCCAGTTATCAACAGTATTAAATTCGATTAAACAAATCATAGTATA
CATAACTCAATCTCACATGAAGTATAGGACAGATCTCAAATAATTGAGAATTTTGCTGAG
AAGTTGTTAGCAAAAAAATATAAATAGATAGCTTTCTTGACAGGAGCAGGAATTAGTGTA
AGTGCTGGAATTCCAGATTTTAGATCTCCTGAAACTGGGCTTTACGCCTAAATAAAAAAA
GAATATGACATAAGTGATCCTTAAAAAATATTTTCAATTAGATATTATCAAGATAATCCT
TTGCCTTTTATGCAAGTTATTAGAGATTTCTTTTCAAGAGAATATCACCCAACTTATGCT
CATAAGTTAATCCATTAAATCTACAAAAGAAAACAACTATTAATAAATATAACCTAAAAT
ATTGATGGCTTAGAGTTAAAAACAGGGATTAATCCCTCAAAAGTAGTCTAAGCACATGGA
CATATGAGAAAGGCTCATTGTGTGAATTGCAATCATATTGTCAATATTGAAACTTACTTA
CAAAACTGCAAACAGTTAAAAAAAACCTAATGCCCCATTTGCAATAATTTAGTGAAGCCA
AAAATTGTTTTCTTTGGAGAGTTTCTTCCTAATGAGTTTTATTAGTCTAGAGATATTTTG
CCAAATTCAGATTGTGTGGTTGTTATGGGAACATCTTTAGGTGTCTTTCCATTTGCTAAT
TTAATAAACGAAGTAGGTACTTCTGTTCCAATATATATAATAAACAATAAACTTCCTAAA
AATATAAGTCATCTTAAAAATCAAATTGAATTTATATAAGGAGATATCAATGAAATTTCT
AAATAAATTATTGATTACATAAATTGA>TTHERM_00112480(gene)
TTGCAAGAAATTTCAAGAAAAGATACCAAATAGTACGCATAATAACTATAAGAAAGTAAT
AAATAACTTTTAAAGAAAGTAGAAACAGATAAATAGGTAGAAAGACTTTTAGAATTAAAT
ACTCAACTTTATCTTTTAATTAAAAGTATTAAATAGTTATTAATTAGTAGAAGAAAATTA
TTTATTTAACAAATATTGCATAAATAATGCATTTAAAAACACTCTTCAAATCATTAATTA
CTCCAAGTAAGTAAATTACTTTGAGAATATCTAACGTATAGCCAGTTATCAACAGTATTA
AATTCGATTAAACAAATCATAGTATACATAACTCAATCTCACATGAAGTATAGGACAGAT
CTCAAATAATTGAGAATTTTGCTGAGAAGTTGTTAGCAAAAAAATATAAATAGATAGCTT
TCTTGACAGGAGCAGGAATTAGTGTAAGTGCTGGAATTCCAGATTTTAGATCTCCTGAAA
CTGGGCTTTACGCCTAAATAAAAAAAGAATATGACATAAGTGATCCTTAAAAAATATTTT
CAATTAGATATTATCAAGATAATCCTTTGCCTTTTATGCAAGTTATTAGAGATTTCTTTT
CAAGAGAATATCACCCAACTTATGCTCATAAGTTAATCCATTAAATCTACAAAAGAAAAC
AACTATTAATAAATATAACCTAAAATATTGATGGCTTAGAGTTAAAAACAGGGATTAATC
CCTCAAAAGTAGTCTAAGCACATGGACATATGAGAAAGGCTCATTGTGTGAATTGCAATC
ATATTGTCAATATTGAAACTTACTTACAAAACTGCAAACAGTTAAAAAAAACCTAATGCC
CCATTTGCAATAATTTAGTGAAGCCAAAAATTGTTTTCTTTGGAGAGTTTCTTCCTAATG
AGTTTTATTAGTCTAGAGATATTTTGCCAAATTCAGATTGTGTGGTTGTTATGGGAACAT
CTTTAGGTGTCTTTCCATTTGCTAATTTAATAAACGAAGTAGGTACTTCTGTTCCAATAT
ATATAATAAACAATAAACTTCCTAAAAATATAAGTCATCTTAAAAATCAAATTGAATTTA
TATAAGGAGATATCAATGAAATTTCTAAATAAATTATTGATTACATAAATTGATATTCAA
AAATATTAATTCCATTATTCAAAAATGAGCATTAAAATATATTTATTTATCTAAATTTAA
ATATCTTAGTTTAAAATAGTTATAATTT>TTHERM_00112480(protein)
MHLKTLFKSLITPSKQITLRISNVQPVINSIKFDQTNHSIHNSISHEVQDRSQIIENFAE
KLLAKKYKQIAFLTGAGISVSAGIPDFRSPETGLYAQIKKEYDISDPQKIFSIRYYQDNP
LPFMQVIRDFFSREYHPTYAHKLIHQIYKRKQLLINITQNIDGLELKTGINPSKVVQAHG
HMRKAHCVNCNHIVNIETYLQNCKQLKKTQCPICNNLVKPKIVFFGEFLPNEFYQSRDIL
PNSDCVVVMGTSLGVFPFANLINEVGTSVPIYIINNKLPKNISHLKNQIEFIQGDINEIS
KQIIDYIN