Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00113040Standard Name
CHS7 (CHitin Synthase)Aliases
PreTt27118 | 8.m00378 | 3812.m02415Description
CHS7 chitin synthase; Chitin synthase family protein; Chitin synthaseGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- chloroplast ribosome (IEA) | GO:0043253
- CURI complex (IEA) | GO:0032545
- extracellular membraneless organelle (IEA) | GO:0043264
- female pronucleus (IEA) | GO:0001939
- M band (IEA) | GO:0031430
- MICOS complex (IEA) | GO:0061617
- obsolete ectoplasm (IEA) | GO:0043265
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-octaprenyl-6-methoxyphenol hydroxylase activity (IEA) | GO:0008681
- 3-hydroxybutyrate dehydrogenase activity (IEA) | GO:0003858
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- adenine nucleotide transmembrane transporter activity (IEA) | GO:0000295
- aliphatic amine oxidase activity (IEA) | GO:0052595
- borate uptake transmembrane transporter activity (IEA) | GO:0080138
- dADP binding (IEA) | GO:0032563
- dAMP binding (IEA) | GO:0032562
- dolichyl pyrophosphate Glc2Man9GlcNAc2 alpha-1,2-glucosyltransferase activity (IEA) | GO:0106073
- ent-kaurene oxidase activity (IEA) | GO:0052615
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- inositol hexakisphosphate 2-phosphatase activity (IEA) | GO:0052826
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- mercury ion transmembrane transporter activity (IEA) | GO:0015097
- mu-type opioid receptor binding (IEA) | GO:0031852
- obsolete 1-chloro-4-nitrosobenzene nitroreductase activity (IEA) | GO:0052603
- obsolete 4-chloronitrobenzene nitroreductase activity (IEA) | GO:0052602
- obsolete aminoacetone:oxygen oxidoreductase(deaminating) activity (IEA) | GO:0052594
- obsolete delta-tocopherol cyclase activity (IEA) | GO:0052604
- obsolete heterotrimeric G-protein (IEA) | GO:0005065
- obsolete protein tagging activity (IEA) | GO:0008638
- obsolete RNA polymerase I transcription regulator recruiting activity (IEA) | GO:0001186
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- RNA polymerase II transcriptional activator activity, copper ion regulated proximal promoter sequence-specific DNA binding (IEA) | GO:0001211
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- transcriptional repressor activity, metal ion regulated sequence-specific DNA binding (IEA) | GO:0001210
- type 1B serotonin receptor binding (IEA) | GO:0031822
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
- uracil oxygenase activity (IEA) | GO:0052614
Biological Process
- apoptotic process involved in tube lumen cavitation (IEA) | GO:0060609
- atrichoblast differentiation (IEA) | GO:0010055
- basement membrane organization (IEA) | GO:0071711
- budding cell bud growth (IEA) | GO:0007117
- budding cell isotropic bud growth (IEA) | GO:0007119
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to dsRNA (IEA) | GO:0071359
- cellular response to iron(III) ion (IEA) | GO:0071283
- cellular response to lead ion (IEA) | GO:0071284
- cellular response to manganese ion (IEA) | GO:0071287
- cytoplasmic actin-based contraction involved in forward cell motility (IEA) | GO:0060328
- establishment of tissue polarity (IEA) | GO:0007164
- extraembryonic membrane development (IEA) | GO:1903867
- free ubiquitin chain depolymerization (IEA) | GO:0010995
- G protein-coupled acetylcholine receptor signaling pathway (IEA) | GO:0007213
- G protein-coupled dopamine receptor signaling pathway (IEA) | GO:0007212
- glycolytic process from fructose through fructose-6-phosphate (IEA) | GO:0061616
- glycolytic process from glycerol (IEA) | GO:0061613
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- high density lipoprotein particle mediated signaling (IEA) | GO:0055097
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- integrin-mediated signaling pathway (IEA) | GO:0007229
- L-alanine import across plasma membrane (IEA) | GO:1904273
- L-ornithine transmembrane transport (IEA) | GO:1903352
- lactate catabolic process (IEA) | GO:1903457
- low-density lipoprotein particle mediated signaling (IEA) | GO:0055096
- maintenance of transcriptional fidelity during DNA-templated transcription elongation from bacterial-type RNA polymerase promoter (IEA) | GO:0001194
- maintenance of transcriptional fidelity during transcription elongation (IEA) | GO:0001192
- maintenance of transcriptional fidelity during transcription elongation by RNA polymerase II (IEA) | GO:0001193
- maternal process involved in female pregnancy (IEA) | GO:0060135
- mesonephric glomerular mesangial cell fate commitment (IEA) | GO:0061264
- mesonephric interstitial fibroblast differentiation (IEA) | GO:0061266
- mitotic nuclear division (IEA) | GO:0007067
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of androst-4-ene-3,17-dione biosynthetic process (IEA) | GO:1903455
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of establishment of Sertoli cell barrier (IEA) | GO:1904445
- negative regulation of interleukin-17-mediated signaling pathway (IEA) | GO:1903882
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phospholipid translocation (IEA) | GO:0061093
- negative regulation of phytoalexin metabolic process (IEA) | GO:0052321
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- Notch receptor processing (IEA) | GO:0007220
- obsolete actin filament polymerization involved in mitotic actomyosin contractile ring assembly (IEA) | GO:1904670
- obsolete detection of organic substance (IEA) | GO:0071703
- obsolete gametophytic self-incompatibility (IEA) | GO:0009872
- obsolete loss of asymmetric budding (IEA) | GO:0007122
- obsolete modulation of catalytic activity in other organism involved in symbiotic interaction (IEA) | GO:0052203
- obsolete negative regulation by host of symbiont defense response (IEA) | GO:0052196
- obsolete negative regulation of transcription during mitotic cell cycle (IEA) | GO:0007068
- obsolete positive regulation of hh target transcription factor activity (IEA) | GO:0007228
- obsolete regulation of histone H3-K79 methylation (IEA) | GO:2001160
- obsolete regulation of mating-type specific transcription from RNA polymerase II promoter (IEA) | GO:0001196
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete reproduction (IEA) | GO:0000003
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- obsolete telomere binding (IEA) | GO:0007003
- ommochrome biosynthetic process (IEA) | GO:0006727
- organic acid transport (IEA) | GO:0015849
- osmosensory signaling pathway (IEA) | GO:0007231
- P granule disassembly (IEA) | GO:1903864
- phagocytosis (IEA) | GO:0006909
- phenanthrene catabolic process via trans-9(S),10(S)-dihydrodiolphenanthrene (IEA) | GO:0018957
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of anterograde dense core granule transport (IEA) | GO:1901953
- positive regulation of ATF6-mediated unfolded protein response (IEA) | GO:1903893
- positive regulation of border follicle cell migration (IEA) | GO:1903688
- positive regulation of forebrain neuron differentiation (IEA) | GO:2000979
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of membrane tubulation (IEA) | GO:1903527
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of transcription of Notch receptor target (IEA) | GO:0007221
- pronephric glomerulus morphogenesis (IEA) | GO:0035775
- pronephric proximal tubule development (IEA) | GO:0035776
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- prostate induction (IEA) | GO:0060514
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of antisense RNA transcription (IEA) | GO:0060194
- regulation of beta-glucan metabolic process (IEA) | GO:0032950
- regulation of border follicle cell migration (IEA) | GO:1903684
- regulation of cellular response to manganese ion (IEA) | GO:1905802
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of dendrite extension (IEA) | GO:1903859
- regulation of heart morphogenesis (IEA) | GO:2000826
- regulation of interleukin-35-mediated signaling pathway (IEA) | GO:0070758
- regulation of metal ion transport (IEA) | GO:0010959
- regulation of microglial cell migration (IEA) | GO:1904139
- regulation of nucleus organization (IEA) | GO:1903353
- regulation of response to type II interferon (IEA) | GO:0060330
- response to high density lipoprotein particle (IEA) | GO:0055099
- response to low-density lipoprotein particle stimulus (IEA) | GO:0055098
- response to oxygen radical (IEA) | GO:0000305
- response to reactive oxygen species (IEA) | GO:0000302
- signal transduction (IEA) | GO:0007165
- vacuolar acidification (IEA) | GO:0007035
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
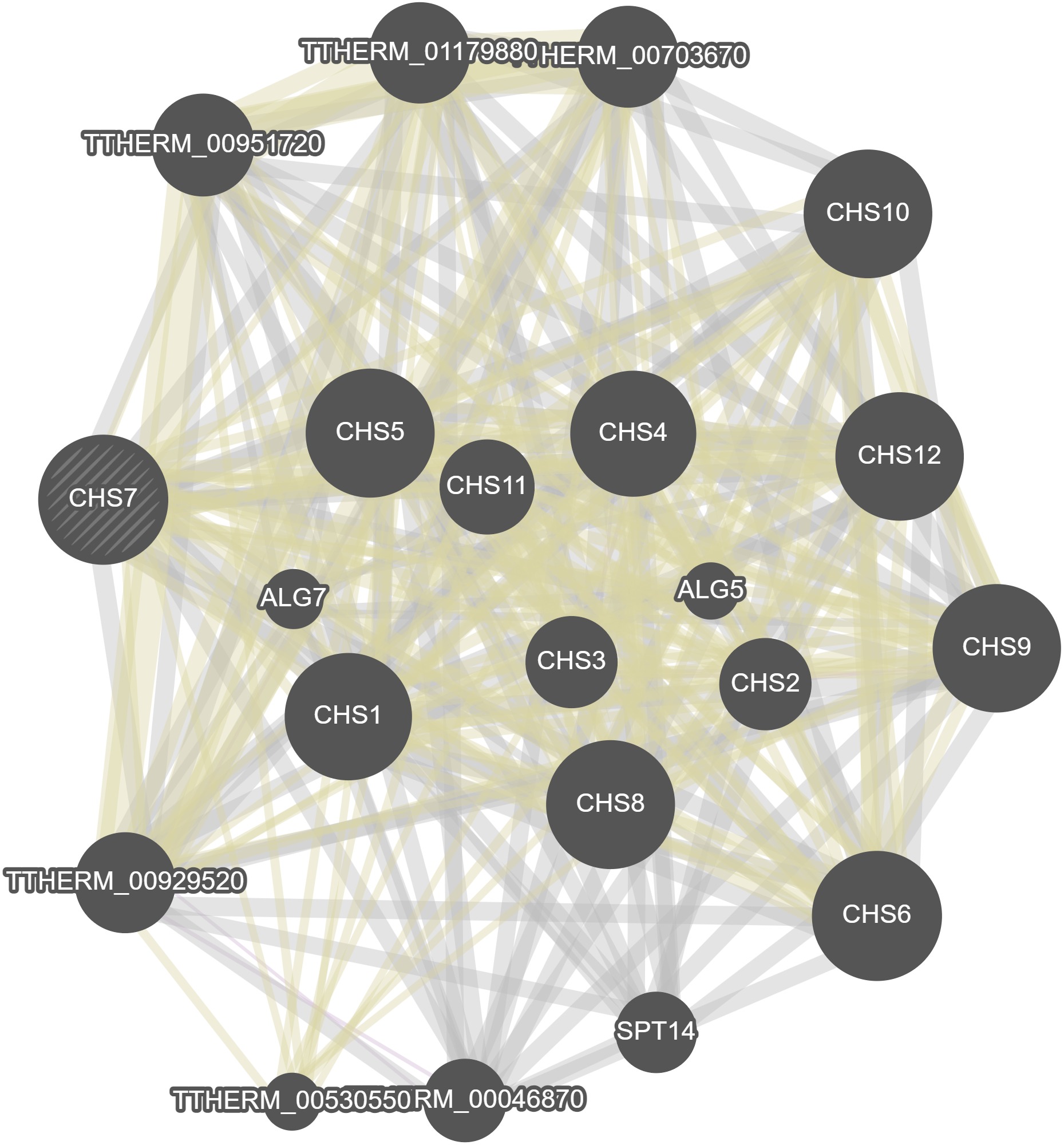
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00113040(coding)
ATGTCAGAAGAATAATCACAATTAAAAAAGAGATTAATTTAAACCGATTTTGAATTAGAT
CAATCTAAAGTTAATGAAAGGCTGATAAACAATACTCCATTATTAGTTGATAACCATTCA
CTATATTAGAAGCCGCTAGACCTTTATATGGTTTGCACAGGTTAAGATGACTTAGATGGG
TAACTAGAACCATTAAACGTTTATACATAAACAATCTAACCAAAGACTCATTACTAATCT
AATGGATAGGCTTATTATACTTACTTTGATGAAACATCTTATAGAAACTAAGGGTTACTA
AGAAATATAAATGAAAAAGATGGGCTGATAGGTTATTATAGCAGTAAAAAGCAAGAAAAT
ATTCTTCAAATGCTTGTTTGTGTCACTATGTATAGTGAAGAGAGAAACTTTCTTGAATAG
ACTCTTCTTCATATACAAAATAATCTCAAAGGATTTTAAGAACTTGAAGTTTAAGATTTT
TAGGTAGTTGTTGCTGTTATCTATGATGGAATCATGAAAGCAAAAGAAGAAGTTGTTGAT
TTTTATCTTGAAATTGATTCCGAAAATGGTTTTGATCTTGATAAAAATTTAAAAAGGAGA
AGAGAAATTATTTAGAGCCAAATGGACTATCTGACATTCACAAATTAATCTCATCTTTTA
GATACAAATGAAGGTCTTCCCCCAACTATACCTAAATAAATATCTTTATTGTATTAAAAT
AGATTTCAATTAAGCCTTCAAGATAAAAGTAATCGCAATTCTAAAGTATAAATAATTTGC
CCTCAAAATCCTCTATTTGACACTAATCATCCTTCCTTAACAGTGTTTTCTCTCTTTAAA
CATAAAAATGCAAAAAAGTTATCTTCTCATTTATGGTTCTTTGAAGGTTTATGTCGCTAA
ACAAGGCCTAAGTATGTTTGCTTTGTGGATGCTGGAACATTACCTTCTTAATATGGTATA
GTCAAATTTTACTAAGCAATGGAGGGAAATTAAACAATAGGAGGAGTATGTGGCTTCTTG
GGATTAGAAGAACCACCTGAAGGAGGAGGAGGAGGAGGAGGAGGAGAAAGTAAATCAGAT
AATGATAAATTAGAAAAATTAAAAAAGAATTTGCAATAATTAAAATAGTAAGATTAAATA
TAATGGGAGTAATAGTATTAATAGGAGAAATAAGATAAATAAAAGGAATTAAGATAAGTC
AAAATTTATACAATGAGTGAAATTAATTAAATTTTTGATCAGTTATCAAGTAACTTCAGA
AATTAAAAGTAGAATCAAATATTCAAAAGAAATTATATTATCACAAACGCAATACTTTTT
TTTATTTTCTTACCCTTTTATTTATTTAAATTTGTACTGATTAATGTTTTATTTAAACTG
ATTTCTAAACTTGGATCTCTTGTAGCCAAATTTTTAGAAAATTTCCTATAAATTTTTGCA
TCTATAAGGCTTGCATAAGTCCAAGAATATGCTATTGGTCATATCATAGATAAAAACTTT
GATTCTTTTTTAGGATTTATGCATGTGCTACCAGGAGCTTGGTCTGCTTACAGATATGAT
GCTCTGGAAGTAAGAAAGGGTGAAAAATCTAATTTTATGCAAGATAGCTACTTTAAATCT
GTTTTGAATCCTGAGCTATTAACAGATGACATCAAAGAAGCAAATAAATTTTTAGCAGAA
GATAGAATACTTTGTCTAGGTATCATTTCAGCAAAAGGATCTAACTACAAATTAAAATAT
ATTCCCGAAGCACTTGCTGTAACAGACTCTCCTGAAACATTAGAAGAATTCATTCACTAG
CACAGAAGATGGACAAATAGCATGATTTTTGCACTCAACCATGTTTTAAAAAATTATAAA
GCCAGTATTTAAGGCAGTAAGCATTCAAATTTTTACAGAAAAATTTGCTTACCAGTTAAT
ATGCTTTTTGCTTATCTTGGGTTAATCAATTATATTGTATTGCCAGCATTTTTCTTGTTT
ATGGTAATATTATCAGGATATTAATTTTACATTCCATCCAACATACATGGAGTTGCTTTT
ATTAATGAAGACCTTGCACAAAATAAATGCCAAATAGATTACCAACTAAAAGAATTTGAT
TTGCAATGTAATTCTGAAATAAAATCAAATTGCTATATAGATTGTTAAACAGGAGAGCTT
AATATTTATCAAAAAATATTTAATATGTTGATGAAAACTTTTCCTTTTGCTATTGTAGTT
ACAGTAGTTGTTTTTATGATAGTATGCCTGTCTTTTAAAATGAAAAAATAAAATCTTTCT
GAGTATTTGAAACTATTTAATTAGGTTTTAACATAAGAGGATAAATAGAAAATATAATCT
TAAAAGCAATAGCTAATAGAAAAATAAAAAACTCCTGAATAAATTGAAACTTGCTCAAAT
AATTTAATTTTTAAGCTTGGAGAAAAAAGATTGAAAGATCTTAAGATTATATTTGATCAA
AACTAATCACATAAACTTCCAAAAGATCTGTAGAAGATAAAATCAGAATATGACAGAATA
TACTCGCAAAAATATTATAACAAAGTTTACAAAATATTTGCTTCTCTCTTTTCAATAGAG
GCTTCAATTTTAATGATCATACTCAGTGCTGCTTTATATATTAATATATTTACTCCAGAT
ACATTCTCTGATCTAGGATTTTTCACACTACCAAAATGGATGAGAGATTATATTATCATA
ATTTTAATATTTAATATGGGGCTTATTTTACTTATTTTATTTTTCCATACAATCTTTTAA
CCAAAGATATTGTGGCTATTTGTAAAATCCTACTTTTCTTATATTTTATACTCTCCAGTG
TATTAAATATACCTAACCATTTTTTCGTTTTGTAATCTTGATGATGTGACATGGGGCACA
AAAGGCTTGCAAAACTCAACACAAAATTAGACATTTTTAACTGACAAAGTTAGATATGTG
AAGTATTGGGTGTTTCTCAACTCTTCTTTAATAATTGCATTCCTTTGTGCTAGCTTGATA
CCATTAGATAAAACTCCATATGTGATTATTGGAATAGGTATATATGGTTCTCTTTACAAC
TCAGTAAGAATTATAGGAGGATCAATAAATTACATAAAATACTTTGTTTTTGATAAAATT
AAAATTAAAAGTGCGTTTAAAAAAATAATATCAGATAATATATAGAAATAAGATGAAATT
AGATATGACTACTAATAAATACTAAATAATTGTAAAAATATTGAATTTAAAATTCAAGAA
CTGAGTGAATCAGAAATAAGCGATGATAATATTTAATTAATAACATCTCGTATGATAAGT
TAATAGTATAGTAATTAAAGCGATTATTAACTCAATATGAATAAGGAACAAAGCTGA>TTHERM_00113040(gene)
TCTATAAAATTTAATTGAAATTGATAATTTTATAGATAATTAATAATTTTGTCATTAATT
AAAAAAAACCAAAAATGTCAGAAGAATAATCACAATTAAAAAAGAGATTAATTTAAACCG
ATTTTGAATTAGATCAATCTAAAGTTAATGAAAGGCTGATAAACAATACTCCATTATTAG
TTGATAACCATTCACTATATTAGAAGCCGCTAGACCTTTATATGGTTTGCACAGGTTAAG
ATGACTTAGATGGGTAACTAGAACCATTAAACGTTTATACATAAACAATCTAACCAAAGA
CTCATTACTAATCTAATGGATAGGCTTATTATACTTACTTTGATGAAACATCTTATAGAA
ACTAAGGGTTACTAAGAAATATAAATGAAAAAGATGGGCTGATAGGTTATTATAGCAGTA
AAAAGCAAGAAAATATTCTTCAAATGCTTGTTTGTGTCACTATGTATAGTGAAGAGAGAA
ACTTTCTTGAATAGACTCTTCTTCATATACAAAATAATCTCAAAGGATTTTAAGAACTTG
AAGTTTAAGATTTTTAGGTAGTTGTTGCTGTTATCTATGATGGAATCATGAAAGCAAAAG
AAGAAGTTGTTGATTTTTATCTTGAAATTGATTCCGAAAATGGTTTTGATCTTGATAAAA
ATTTAAAAAGGAGAAGAGAAATTATTTAGAGCCAAATGGACTATCTGACATTCACAAATT
AATCTCATCTTTTAGATACAAATGAAGGTCTTCCCCCAACTATACCTAAATAAATATCTT
TATTGTATTAAAATAGATTTCAATTAAGCCTTCAAGATAAAAGTAATCGCAATTCTAAAG
TATAAATAATTTGCCCTCAAAATCCTCTATTTGACACTAATCATCCTTCCTTAACAGTGT
TTTCTCTCTTTAAACATAAAAATGCAAAAAAGTTATCTTCTCATTTATGGTTCTTTGAAG
GTTTATGTCGCTAAACAAGGCCTAAGTATGTTTGCTTTGTGGATGCTGGAACATTACCTT
CTTAATATGGTATAGTCAAATTTTACTAAGCAATGGAGGGAAATTAAACAATAGGAGGAG
TATGTGGCTTCTTGGGATTAGAAGAACCACCTGAAGGAGGAGGAGGAGGAGGAGGAGGAG
AAAGTAAATCAGATAATGATAAATTAGAAAAATTAAAAAAGAATTTGCAATAATTAAAAT
AGTAAGATTAAATATAATGGGAGTAATAGTATTAATAGGAGAAATAAGATAAATAAAAGG
AATTAAGATAAGTCAAAATTTATACAATGAGTGAAATTAATTAAATTTTTGATCAGTTAT
CAAGTAACTTCAGAAATTAAAAGTAGAATCAAATATTCAAAAGAAATTATATTATCACAA
ACGCAATACTTTTTTTTATTTTCTTACCCTTTTATTTATTTAAATTTGTACTGATTAATG
TTTTATTTAAACTGATTTCTAAACTTGGATCTCTTGTAGCCAAATTTTTAGAAAATTTCC
TATAAATTTTTGCATCTATAAGGCTTGCATAAGTCCAAGAATATGCTATTGGTCATATCA
TAGATAAAAACTTTGATTCTTTTTTAGGATTTATGCATGTGCTACCAGGAGCTTGGTCTG
CTTACAGATATGATGCTCTGGAAGTAAGAAAGGGTGAAAAATCTAATTTTATGCAAGATA
GCTACTTTAAATCTGTTTTGAATCCTGAGCTATTAACAGATGACATCAAAGAAGCAAATA
AATTTTTAGCAGAAGATAGAATACTTTGTCTAGGTATCATTTCAGCAAAAGGATCTAACT
ACAAATTAAAATATATTCCCGAAGCACTTGCTGTAACAGACTCTCCTGAAACATTAGAAG
AATTCATTCACTAGCACAGAAGATGGACAAATAGCATGATTTTTGCACTCAACCATGTTT
TAAAAAATTATAAAGCCAGTATTTAAGGCAGTAAGCATTCAAATTTTTACAGAAAAATTT
GCTTACCAGTTAATATGCTTTTTGCTTATCTTGGGTTAATCAATTATATTGTATTGCCAG
CATTTTTCTTGTTTATGGTAATATTATCAGGATATTAATTTTACATTCCATCCAACATAC
ATGGAGTTGCTTTTATTAATGAAGACCTTGCACAAAATAAATGCCAAATAGATTACCAAC
TAAAAGAATTTGATTTGCAATGTAATTCTGAAATAAAATCAAATTGCTATATAGATTGTT
AAACAGGAGAGCTTAATATTTATCAAAAAATATTTAATATGTTGATGAAAACTTTTCCTT
TTGCTATTGTAGTTACAGTAGTTGTTTTTATGATAGTATGCCTGTCTTTTAAAATGAAAA
AATAAAATCTTTCTGAGTATTTGAAACTATTTAATTAGGTTTTAACATAAGAGGATAAAT
AGAAAATATAATCTTAAAAGCAATAGCTAATAGAAAAATAAAAAACTCCTGAATAAATTG
AAACTTGCTCAAATAATTTAATTTTTAAGCTTGGAGAAAAAAGATTGAAAGATCTTAAGA
TTATATTTGATCAAAACTAATCACATAAACTTCCAAAAGATCTGTAGAAGATAAAATCAG
AATATGACAGAATATACTCGCAAAAATATTATAACAAAGTTTACAAAATATTTGCTTCTC
TCTTTTCAATAGAGGCTTCAATTTTAATGATCATACTCAGTGCTGCTTTATATATTAATA
TATTTACTCCAGATACATTCTCTGATCTAGGATTTTTCACACTACCAAAATGGATGAGAG
ATTATATTATCATAATTTTAATATTTAATATGGGGCTTATTTTACTTATTTTATTTTTCC
ATACAATCTTTTAACCAAAGATATTGTGGCTATTTGTAAAATCCTACTTTTCTTATATTT
TATACTCTCCAGTGTATTAAATATACCTAACCATTTTTTCGTTTTGTAATCTTGATGATG
TGACATGGGGCACAAAAGGCTTGCAAAACTCAACACAAAATTAGACATTTTTAACTGACA
AAGTTAGATATGTGAAGTATTGGGTGTTTCTCAACTCTTCTTTAATAATTGCATTCCTTT
GTGCTAGCTTGATACCATTAGATAAAACTCCATATGTGATTATTGGAATAGGTATATATG
GTTCTCTTTACAACTCAGTAAGAATTATAGGAGGATCAATAAATTACATAAAATACTTTG
TTTTTGATAAAATTAAAATTAAAAGTGCGTTTAAAAAAATAATATCAGATAATATATAGA
AATAAGATGAAATTAGATATGACTACTAATAAATACTAAATAATTGTAAAAATATTGAAT
TTAAAATTCAAGAACTGAGTGAATCAGAAATAAGCGATGATAATATTTAATTAATAACAT
CTCGTATGATAAGTTAATAGTATAGTAATTAAAGCGATTATTAACTCAATATGAATAAGG
AACAAAGCTGAATTAATAACATACAAAAATAAAAGAATGAAAAATAATATTTTAGTATGG
TTAATAATATAAATTTATAC>TTHERM_00113040(protein)
MSEEQSQLKKRLIQTDFELDQSKVNERLINNTPLLVDNHSLYQKPLDLYMVCTGQDDLDG
QLEPLNVYTQTIQPKTHYQSNGQAYYTYFDETSYRNQGLLRNINEKDGLIGYYSSKKQEN
ILQMLVCVTMYSEERNFLEQTLLHIQNNLKGFQELEVQDFQVVVAVIYDGIMKAKEEVVD
FYLEIDSENGFDLDKNLKRRREIIQSQMDYLTFTNQSHLLDTNEGLPPTIPKQISLLYQN
RFQLSLQDKSNRNSKVQIICPQNPLFDTNHPSLTVFSLFKHKNAKKLSSHLWFFEGLCRQ
TRPKYVCFVDAGTLPSQYGIVKFYQAMEGNQTIGGVCGFLGLEEPPEGGGGGGGGESKSD
NDKLEKLKKNLQQLKQQDQIQWEQQYQQEKQDKQKELRQVKIYTMSEINQIFDQLSSNFR
NQKQNQIFKRNYIITNAILFFIFLPFYLFKFVLINVLFKLISKLGSLVAKFLENFLQIFA
SIRLAQVQEYAIGHIIDKNFDSFLGFMHVLPGAWSAYRYDALEVRKGEKSNFMQDSYFKS
VLNPELLTDDIKEANKFLAEDRILCLGIISAKGSNYKLKYIPEALAVTDSPETLEEFIHQ
HRRWTNSMIFALNHVLKNYKASIQGSKHSNFYRKICLPVNMLFAYLGLINYIVLPAFFLF
MVILSGYQFYIPSNIHGVAFINEDLAQNKCQIDYQLKEFDLQCNSEIKSNCYIDCQTGEL
NIYQKIFNMLMKTFPFAIVVTVVVFMIVCLSFKMKKQNLSEYLKLFNQVLTQEDKQKIQS
QKQQLIEKQKTPEQIETCSNNLIFKLGEKRLKDLKIIFDQNQSHKLPKDLQKIKSEYDRI
YSQKYYNKVYKIFASLFSIEASILMIILSAALYINIFTPDTFSDLGFFTLPKWMRDYIII
ILIFNMGLILLILFFHTIFQPKILWLFVKSYFSYILYSPVYQIYLTIFSFCNLDDVTWGT
KGLQNSTQNQTFLTDKVRYVKYWVFLNSSLIIAFLCASLIPLDKTPYVIIGIGIYGSLYN
SVRIIGGSINYIKYFVFDKIKIKSAFKKIISDNIQKQDEIRYDYQQILNNCKNIEFKIQE
LSESEISDDNIQLITSRMISQQYSNQSDYQLNMNKEQS