Gene Model Identifier
TTHERM_00129830
Standard Name
HEX1
(beta-HEXosaminidase )
Aliases
PreTt09534 | 9.m00555 | 3699.m00263
Description
HEX1 glycosyl hydrolase family 20 beta-N-acetylhexosaminidase family protein; Lysosomal beta-hexosaminidase; synthesized as a precursor that is processed into several mature glycosylated isozymes; isozymes are secreted during both growth and starvation; a fraction remains covalently bound to the plasma membrane; Beta-hexosaminidase
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
>TTHERM_00129830(coding)
ATGCAAAAGATACTTTTAATTACTTTCCTTCTTGGAATAGCTCTCGCTCAAATTACTCCT
GGCGTTGACCCTATTTCAGCTAAGGTTATGCCTAAACCTAAGAATTACACTTATGGAGAT
TTGAGCTTACTTGTCACTGATCCTTGCGGAATCTCTTACAGACCTTCTGTTGGGTCAGGA
AAAGTACCCAACCATGTCTATCAAATTATTGGATTCTACACTTTGAATATTTTCAATTCT
AACGAAAACTCTTGTGCTATGTAAAGAGAATTGTATAAGAATGAAACAACCATTGAAAAG
ATGCGTAGATTACAACATTCCTAAAATATAGTCTTCGATATTTTTATCTAAGACGCTGCT
TTGGCCACTGCAGACACACTCGAAGACGAATATTATGATTTATAAATTTATAATACCACA
TATTGGAAATTGACTGCTAACAAATATGTTGGTTTACTCCGTGGTTTAGAAACTTACTCT
CAATTATTCACTTAAGACGAAGACACTGAAGATTGGTATTTGAATAACATCCCTATTTCT
ATTCAAGATTAACCTGACTACATCTACAGAGGTCTTATGATAGATTCAGCCAGACATTTC
TTATCAGTTGAAACTATTTTAAAAACTATTGATTCTATGTTATTCAACAAGTTGAATGTT
CTCCATTGGCACATCACTGATACTGAATCCTTCCCCTTCCCTCTTAAATCATTCCCTAAT
ATTACTAAATATGGAGCCTACTCTAAGAAGAAACAATACAGCTTCGAAGACATTTAATAC
ATTGTAGACTAAGCTCTCAACAAGGGTATTTAAGTTATTCCTGAAGTCGATTCTCCAGGA
CACGCTTTTTCATGGGCTAGATCTCCTTAATTCTCTAGTATTGGTCTATTATGTGATTAA
TATAATGGATAGTTAGACCCAACACTAAATTTAACTTACACTGCTGTTAAGGGTATTATG
GAAGATATGAATACTTAATTCTACACTGCTAAGTATGTTCATTTTGGTGGTGATGAAGTT
GAAGAATAATGCTGGAATAAACGCCCTGAAATTAAGGAATTCATGAATTAAAATAACATC
TCTACATATACTGATTTGTAGAATTATTACAGAAAGAACTAAGTTAACATTTGGAAATCA
ATTAATGCTACTAAGCCTGCTATTTTCTGGGCAGATTCAAATACTTTGAAATATGGTCCT
GATGATATTATTCAATGGTGGGGATCTACTCATGATTTTTCTTCAATCAAAGATCTTCCT
AACAAAATAATTTTATCTTTCTATGATAATACTTATTTGGATGTTGGTGAGGGAAATAGA
TATGGTGGAAGTTATGGCAGCATGTATAACTGGGATGTCTTAAACTCTTTCAATCCTAGA
GTTCCTGGAATTAAGGGTGAAATTCTTGGTGGCGAAACATGCTTATGGAGTGAAATGAAT
GATGATTCTACTTAATTCTAAAGACTTTGGACAAGAAATAGTGCATTTGCTGAAAGACTT
TGGAACACTGACGCTGCTAATAATGAAACTTACAAAACTAGAGCTTTAGTTAGCAGAATG
GTCTTTATGCAACACCGTTTAACTGCTAGAGGAATCCCTGCTTCTCCTGTAACAGTTGGT
ATTTGTGAATAAAACCTTTCTCTCTGCTACAATTGA
>TTHERM_00129830(gene)
CCCTTAATAATTTTAATATATATATTAAAAATACACTGATTTAATTACATAATAAATTAG
CAGTAATAAAAAATTCTAAATATATTGATTGTAGCTATGCAAAAGATACTTTTAATTACT
TTCCTTCTTGGAATAGCTCTCGCTCAAATTACTCCTGGCGTTGACCCTATTTCAGCTAAG
GTTATGCCTAAACCTAAGAATTACACTTATGGAGATTTGAGCTTACTTGTCACTGATCCT
TGCGGAATCTCTTACAGACCTTCTGTTGGGTCAGGAAAAGTACCCAACCATGTCTATCAA
ATTATTGGATTCTACACTTTGAATATTTTCAATTCTAACGAAAACTCTTGTGCTATGTAA
AGAGAATTGTATAAGAATGAAACAACCATTGAAAAGATGCGTAGATTACAACATTCCTAA
AATATAGTCTTCGATATTTTTATCTAAGACGCTGCTTTGGCCACTGCAGACACACTCGAA
GACGAATATTATGATTTATAAATTTATAATACCACATATTGGAAATTGACTGCTAACAAA
TATGTTGGTTTACTCCGTGGTTTAGAAACTTACTCTCAATTATTCACTTAAGACGAAGAC
ACTGAAGATTGGTATTTGAATAACATCCCTATTTCTATTCAAGATTAACCTGACTACATC
TACAGAGGTCTTATGATAGATTCAGCCAGACATTTCTTATCAGTTGAAACTATTTTAAAA
ACTATTGATTCTATGTTATTCAACAAGTTGAATGTTCTCCATTGGCACATCACTGATACT
GAATCCTTCCCCTTCCCTCTTAAATCATTCCCTAATATTACTAAATATGGAGCCTACTCT
AAGAAGAAACAATACAGCTTCGAAGACATTTAATACATTGTAGACTAAGCTCTCAACAAG
GGTATTTAAGTTATTCCTGAAGTCGATTCTCCAGGACACGCTTTTTCATGGGCTAGATCT
CCTTAATTCTCTAGTATTGGTCTATTATGTGATTAATATAATGGATAGTTAGACCCAACA
CTAAATTTAACTTACACTGCTGTTAAGGGTATTATGGAAGATATGAATACTTAATTCTAC
ACTGCTAAGTATGTTCATTTTGGTGGTGATGAAGTTGAAGAATAATGCTGGAATAAACGC
CCTGAAATTAAGGAATTCATGAATTAAAATAACATCTCTACATATACTGATTTGTAGAAT
TATTACAGAAAGAACTAAGTTAACATTTGGAAATCAATTAATGCTACTAAGCCTGCTATT
TTCTGGGCAGATTCAAATACTTTGAAATATGGTCCTGATGATATTATTCAATGGTGGGGA
TCTACTCATGATTTTTCTTCAATCAAAGATCTTCCTAACAAAATAATTTTATCTTTCTAT
GATAATACTTATTTGGATGTTGGTGAGGGAAATAGATATGGTGGAAGTTATGGCAGCATG
TATAACTGGGATGTCTTAAACTCTTTCAATCCTAGAGTTCCTGGAATTAAGGGTGAAATT
CTTGGTGGCGAAACATGCTTATGGAGTGAAATGAATGATGATTCTACTTAATTCTAAAGA
CTTTGGACAAGAAATAGTGCATTTGCTGAAAGACTTTGGAACACTGACGCTGCTAATAAT
GAAACTTACAAAACTAGAGCTTTAGTTAGCAGAATGGTCTTTATGCAACACCGTTTAACT
GCTAGAGGAATCCCTGCTTCTCCTGTAACAGTTGGTATTTGTGAATAAAACCTTTCTCTC
TGCTACAATTGATTCTAAATATAAAAATTAAATAAATATTTTAAGAAATATTTTTAAGAA
TATTTTAGTATAAAAACTGTATTTTAATTGATAAAAAAAATATAAATATTATTATTAATT
GAATTTTAGCTATTAATTTTATTAATATCTGAAACATATCTTAATTTAATTTTACTAAGT
GTTCATCCCTTTTTTAATTTAGCATAGAGATGGTCACATATTTTATTTTTTTAAAAATGT
ATGTACTTTTTTAAATTAGAAGTTTTTAAAAAATTATGGTATTTAATTCTTTAATTTATT
TCAAATATGAGTTTAAAA
>TTHERM_00129830(protein)
MQKILLITFLLGIALAQITPGVDPISAKVMPKPKNYTYGDLSLLVTDPCGISYRPSVGSG
KVPNHVYQIIGFYTLNIFNSNENSCAMQRELYKNETTIEKMRRLQHSQNIVFDIFIQDAA
LATADTLEDEYYDLQIYNTTYWKLTANKYVGLLRGLETYSQLFTQDEDTEDWYLNNIPIS
IQDQPDYIYRGLMIDSARHFLSVETILKTIDSMLFNKLNVLHWHITDTESFPFPLKSFPN
ITKYGAYSKKKQYSFEDIQYIVDQALNKGIQVIPEVDSPGHAFSWARSPQFSSIGLLCDQ
YNGQLDPTLNLTYTAVKGIMEDMNTQFYTAKYVHFGGDEVEEQCWNKRPEIKEFMNQNNI
STYTDLQNYYRKNQVNIWKSINATKPAIFWADSNTLKYGPDDIIQWWGSTHDFSSIKDLP
NKIILSFYDNTYLDVGEGNRYGGSYGSMYNWDVLNSFNPRVPGIKGEILGGETCLWSEMN
DDSTQFQRLWTRNSAFAERLWNTDAANNETYKTRALVSRMVFMQHRLTARGIPASPVTVG
ICEQNLSLCYN
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
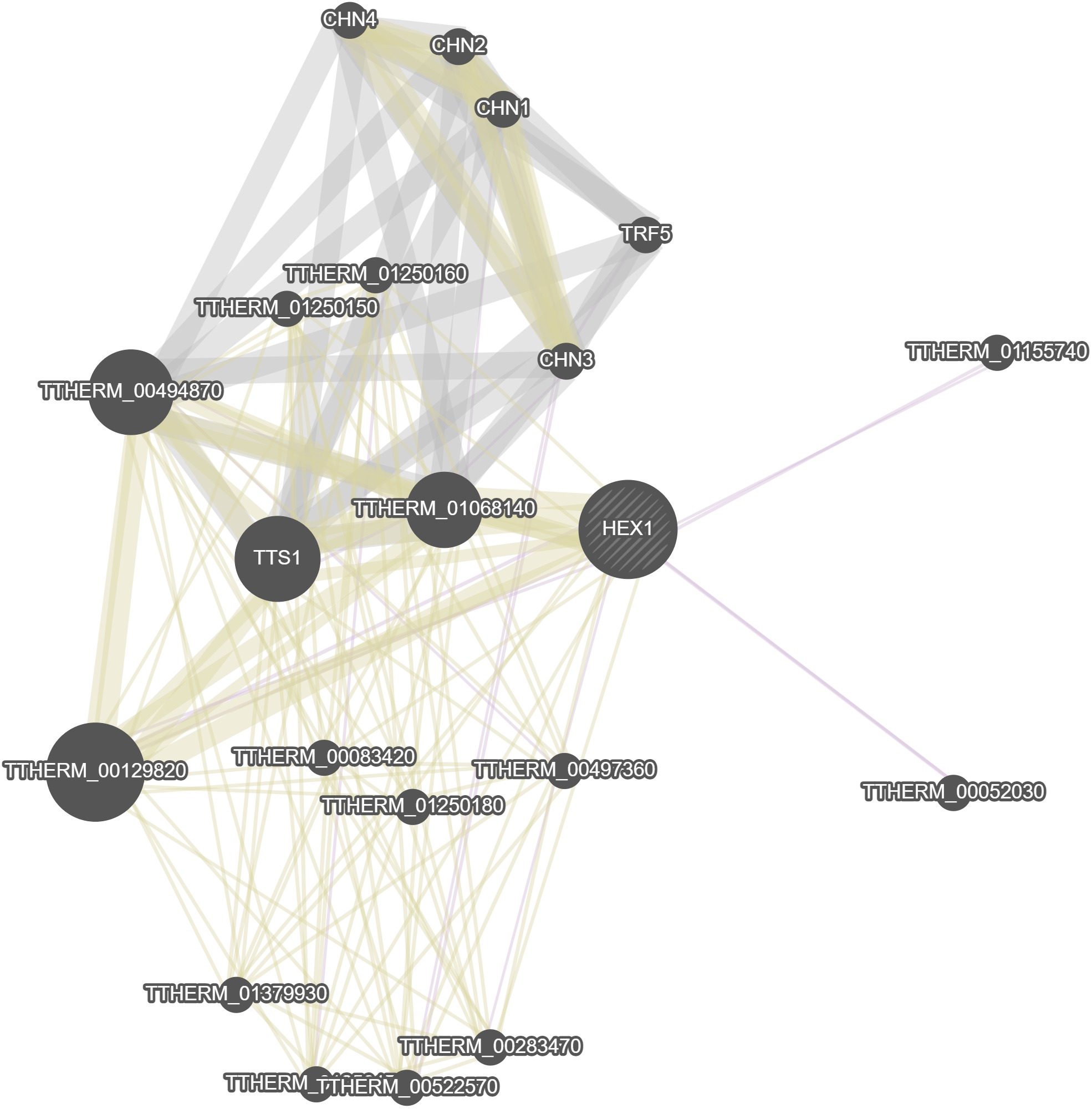
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences