Gene Model Identifier
TTHERM_00146040
Standard Name
Brop-3
(Bromodomain containing protein 3)
Aliases
PreTt26811 | 11.m00508 | 3702.m00230
Description
Brop-3 bromodomain protein; Bromodomain-containing chromatin reader
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
>TTHERM_00146040(coding)
ATGAGCAAAGGCAAGAAAGATAAATTAGATTTAACAGTGGCATTAAATAATGTACTAAAC
GATATAATAGTTCAGGATCCTTAGCAAATATTTTATCTTCCTGTTGATACTAAGCTGTTT
TTTGATTATACAAAGAAAATAGCAAAGCCTATGGACTTATAAACCATGCAAAAAAAAATT
AAAAAGTATGTAGGCTATAAAGAATTTGAAAACGATCTTAATCTGATTTGGAGTAATTGT
AGAATCTACAATTAAGAAAATAGTGACATATATAAAGCATCTTTAAGATTGGAAAGATTA
TCAAAAAAGTCATTAAAGGAGCATCTTCTACCTTTTAAAGCTCATTATAGATTTGATGAG
AATGGAGTTCCAATTAAAAAAAGAAAATACAATCGTGTGAAGGATTCGTTAAACTCAAGT
TTGATTTAAACAAATGGAAATTAAAACTATTAATATGCATCATCTTATGGTATTCCTTCC
AGAAAAACATCCTTAAGTGTAGATAAAAAAGAAAAAAGAGGCTCTAAAGGAGTAGCAAAA
TTGGCAAAGGCTATAAAGAAAGGGAAAAAGCAAGATTTGTTAGGATTAAAAAAAGCTCAA
TAAATACCAGCTCTCCCTTAGAATTTAATATTTGACAAAAAAACTATAATTCACCCTCCT
AAATCTTCAAATTACAACTGTATTGTGATAGGATTGCAATAATAAATCTAATCTAAAGAA
AATCTTATTACTGAAGATGATAAGTAATTTTTGAAGTAGAAGGTTGTGAATTTTTCGTAA
GAAGAACTCTTTAAATGCATTAAAACAATATAAGAATATGAGCCTTAGCACATTTCCTAG
CAATCAAAAGAACACTATATTGTTAATTTTAATACACTCACTAAAATGGTTTTTAATAAA
ATAAAAGAATAATTTTCATTATGA
>TTHERM_00146040(gene)
TTCCCACAAACTCATCTGGCAAGAGAAGAATTTTTCTACATAGAAATTATAATTCATTTT
TTCTCTCTTACTTTACTTTTTACATAAATTGGTTGTGTGAAATACTTAAATAAAAATGAG
CAAAGGCAAGAAAGATAAATTAGATTTAACAGTGGCATTAAATAATGTACTAAACGATAT
AATAGTTCAGGATCCTTAGCAAATATTTTATCTTCCTGTTGATACTAAGCTGTTTTTTGA
TTATACAAAGAAAATAGCAAAGCCTATGGACTTATAAACCATGCAAAAAAAAATTAAAAA
GTATGTAGGCTATAAAGAATTTGAAAACGATCTTAATCTGATTTGGAGTAATTGTAGAAT
CTACAATTAAGAAAATAGTGACATATATAAAGCATCTTTAAGATTGGAAAGATTATCAAA
AAAGTCATTAAAGGAGCATCTTCTACCTTTTAAAGCTCATTATAGATTTGATGAGAATGG
AGTTCCAATTAAAAAAAGAAAATACAATCGTGTGAAGGATTCGTTAAACTCAAGTTTGAT
TTAAACAAATGGAAATTAAAACTATTAATATGCATCATCTTATGGTATTCCTTCCAGAAA
AACATCCTTAAGTGTAGATAAAAAAGAAAAAAGAGGCTCTAAAGGAGTAGCAAAATTGGC
AAAGGCTATAAAGAAAGGGAAAAAGCAAGATTTGTTAGGATTAAAAAAAGCTCAATAAAT
ACCAGCTCTCCCTTAGAATTTAATATTTGACAAAAAAACTATAATTCACCCTCCTAAATC
TTCAAATTACAACTGTATTGTGATAGGATTGCAATAATAAATCTAATCTAAAGAAAATCT
TATTACTGAAGATGATAAGTAATTTTTGAAGTAGAAGGTTGTGAATTTTTCGTAAGAAGA
ACTCTTTAAATGCATTAAAACAATATAAGAATATGAGCCTTAGCACATTTCCTAGCAATC
AAAAGAACACTATATTGTTAATTTTAATACACTCACTAAAATGGTTTTTAATAAAATAAA
AGAATAATTTTCATTATGAAAAACTAACTTTTTTCTGGGAAATAGACAATAAATAAATAA
ATAAGTATTTAATCTAAAAATGAATTTTATACTCTAAATTAAGTAATCTGATTAGTAAAT
TTAAATGATAAAAATTGTTAAATATGTAATACATATCATTTGTAGTAATATTTTTTGCTA
ATTTTGGAAAATAATTGC
>TTHERM_00146040(protein)
MSKGKKDKLDLTVALNNVLNDIIVQDPQQIFYLPVDTKLFFDYTKKIAKPMDLQTMQKKI
KKYVGYKEFENDLNLIWSNCRIYNQENSDIYKASLRLERLSKKSLKEHLLPFKAHYRFDE
NGVPIKKRKYNRVKDSLNSSLIQTNGNQNYQYASSYGIPSRKTSLSVDKKEKRGSKGVAK
LAKAIKKGKKQDLLGLKKAQQIPALPQNLIFDKKTIIHPPKSSNYNCIVIGLQQQIQSKE
NLITEDDKQFLKQKVVNFSQEELFKCIKTIQEYEPQHISQQSKEHYIVNFNTLTKMVFNK
IKEQFSL
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
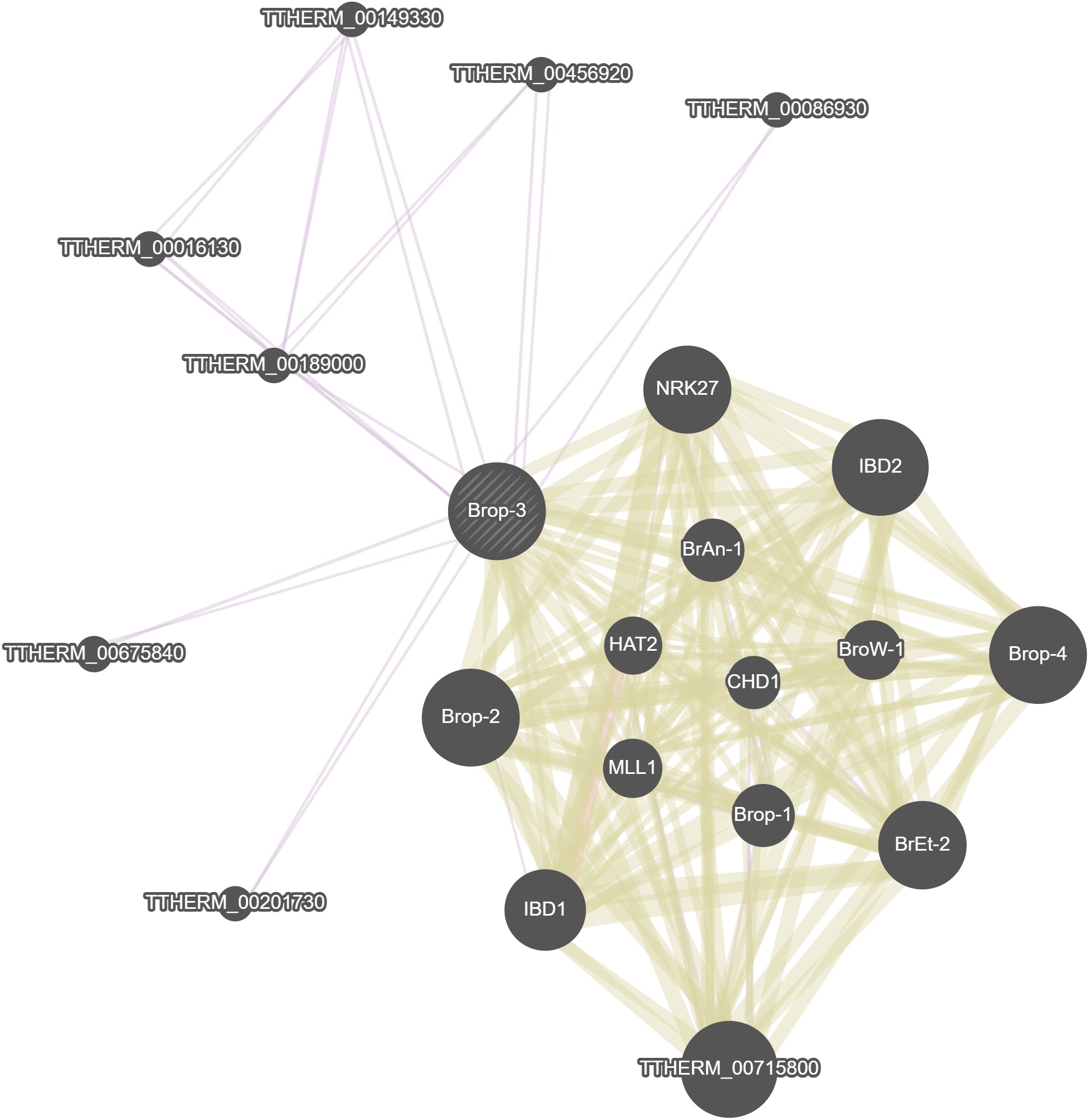
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences