Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00207260Standard Name
RAB45 (RAB GTPase 45)Aliases
PreTt13802 | 17.m00409Description
RAB45 Ras family protein; Novel Rab family protein; small monomeric GTPase that regulates membrane fusion and fission events; Small GTPaseGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- dense core granule membrane (IEA) | GO:0032127
- glutathione hydrolase complex (IEA) | GO:0061672
- heterochromatin domain (IEA) | GO:1990343
- obsolete integral component of mitochondrial membrane (IEA) | GO:0032592
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- alpha-humulene synthase activity (IEA) | GO:0080017
- archaeosine synthase activity (IEA) | GO:0002948
- Atg12 ligase activity (IEA) | GO:0061660
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- FAT10 ligase activity (IEA) | GO:0061661
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- glucose 6-phosphate:phosphate antiporter activity (IEA) | GO:0061513
- growth hormone-releasing hormone receptor activity (IEA) | GO:0016520
- insulin-like growth factor II binding (IEA) | GO:0031995
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- lipopolysaccharide heptosyltransferase activity (IEA) | GO:0008920
- obsolete 3-HSA hydroxylase activity (IEA) | GO:0034819
- obsolete aspartic-type signal peptidase activity (IEA) | GO:0009049
- obsolete autolysin activity (IEA) | GO:0015466
- obsolete citronellol dehydrogenase activity (IEA) | GO:0034821
- obsolete peroxisome-assembly ATPase activity (IEA) | GO:0008573
- obsolete serpin (IEA) | GO:0004868
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- oxidoreductase activity, acting on CH or CH2 groups, with an iron-sulfur protein as acceptor (IEA) | GO:0052592
- phosphatidylinositol-3-phosphate binding (IEA) | GO:0032266
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- Pup ligase activity (IEA) | GO:0061664
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- tetrahydrofolyl-poly(glutamate) polymer binding (IEA) | GO:1904493
- transforming growth factor beta receptor activity, type III (IEA) | GO:0070123
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
- UFM1 ligase activity (IEA) | GO:0061666
Biological Process
- cellular response to vanadate(3-) (IEA) | GO:1902439
- centriole elongation (IEA) | GO:0061511
- dCMP phosphorylation (IEA) | GO:0061567
- detection of stimulus involved in Dma1-dependent checkpoint (IEA) | GO:1902419
- dichotomous subdivision of terminal units involved in lung branching (IEA) | GO:0060448
- embryonic brain development (IEA) | GO:1990403
- extraembryonic membrane development (IEA) | GO:1903867
- ferrous iron transmembrane transport (IEA) | GO:1903874
- flocculation via extracellular polymer (IEA) | GO:0032128
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- glycolytic process from glycerol (IEA) | GO:0061613
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor (IEA) | GO:0002416
- immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor (IEA) | GO:0002415
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- interleukin-19 production (IEA) | GO:0032622
- interleukin-2 production (IEA) | GO:0032623
- L-alanine catabolic process (IEA) | GO:0042853
- mitotic cytokinetic cell separation (IEA) | GO:1902409
- mitotic nuclear division (IEA) | GO:0007067
- modulation by organism of innate immune response in other organism involved in symbiotic interaction (IEA) | GO:0052306
- negative regulation by organism of defense-related jasmonic acid-mediated signal transduction pathway in other organism involved in symbiotic interaction (IEA) | GO:0052267
- negative regulation of calcium ion binding (IEA) | GO:1901877
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of peripheral B cell anergy (IEA) | GO:0002918
- negative regulation of peripheral B cell deletion (IEA) | GO:0002909
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phospholipid translocation (IEA) | GO:0061093
- negative regulation of phytoalexin metabolic process (IEA) | GO:0052321
- negative regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process (IEA) | GO:0062026
- negative regulation of vasculogenesis (IEA) | GO:2001213
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete activation of MAPK activity (IEA) | GO:0000187
- obsolete cellular response to ouabain (IEA) | GO:1905105
- obsolete modulation of peptidase activity in other organism involved in symbiotic interaction (IEA) | GO:0052198
- obsolete multi-organism cellular localization (IEA) | GO:1902581
- obsolete negative regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity (IEA) | GO:1904218
- obsolete positive regulation by host of symbiont defense response (IEA) | GO:0052197
- obsolete positive regulation of nucleic acid-templated transcription (IEA) | GO:1903508
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- phenanthrene catabolic process via trans-9(R),10(R)-dihydrodiolphenanthrene (IEA) | GO:0018956
- plant-type cell wall modification (IEA) | GO:0009827
- polyamine acetylation (IEA) | GO:0032917
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of actin binding (IEA) | GO:1904618
- positive regulation of antifungal innate immune response (IEA) | GO:1905036
- positive regulation of ATF6-mediated unfolded protein response (IEA) | GO:1903893
- positive regulation of deacetylase activity (IEA) | GO:0090045
- positive regulation of fatty acid beta-oxidation (IEA) | GO:0032000
- positive regulation of I-kappaB phosphorylation (IEA) | GO:1903721
- positive regulation of peripheral B cell anergy (IEA) | GO:0002919
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- prostate induction (IEA) | GO:0060514
- protein localization to astral microtubule (IEA) | GO:1902888
- protein localization to cilium (IEA) | GO:0061512
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of antimicrobial peptide production (IEA) | GO:0002784
- regulation of antisense RNA transcription (IEA) | GO:0060194
- regulation of BMP secretion (IEA) | GO:2001284
- regulation of chitin metabolic process (IEA) | GO:0032882
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of dendrite extension (IEA) | GO:1903859
- regulation of PERK-mediated unfolded protein response (IEA) | GO:1903897
- regulation of phosphatidylcholine biosynthetic process (IEA) | GO:2001245
- regulation of striated muscle contraction (IEA) | GO:0006942
- single-organism intracellular transport (IEA) | GO:1902582
- striated muscle contraction (IEA) | GO:0006941
- tRNA threonylcarbamoyladenosine modification (IEA) | GO:0002949
 Domains
Domains
- ( PF00071 ) Ras family
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
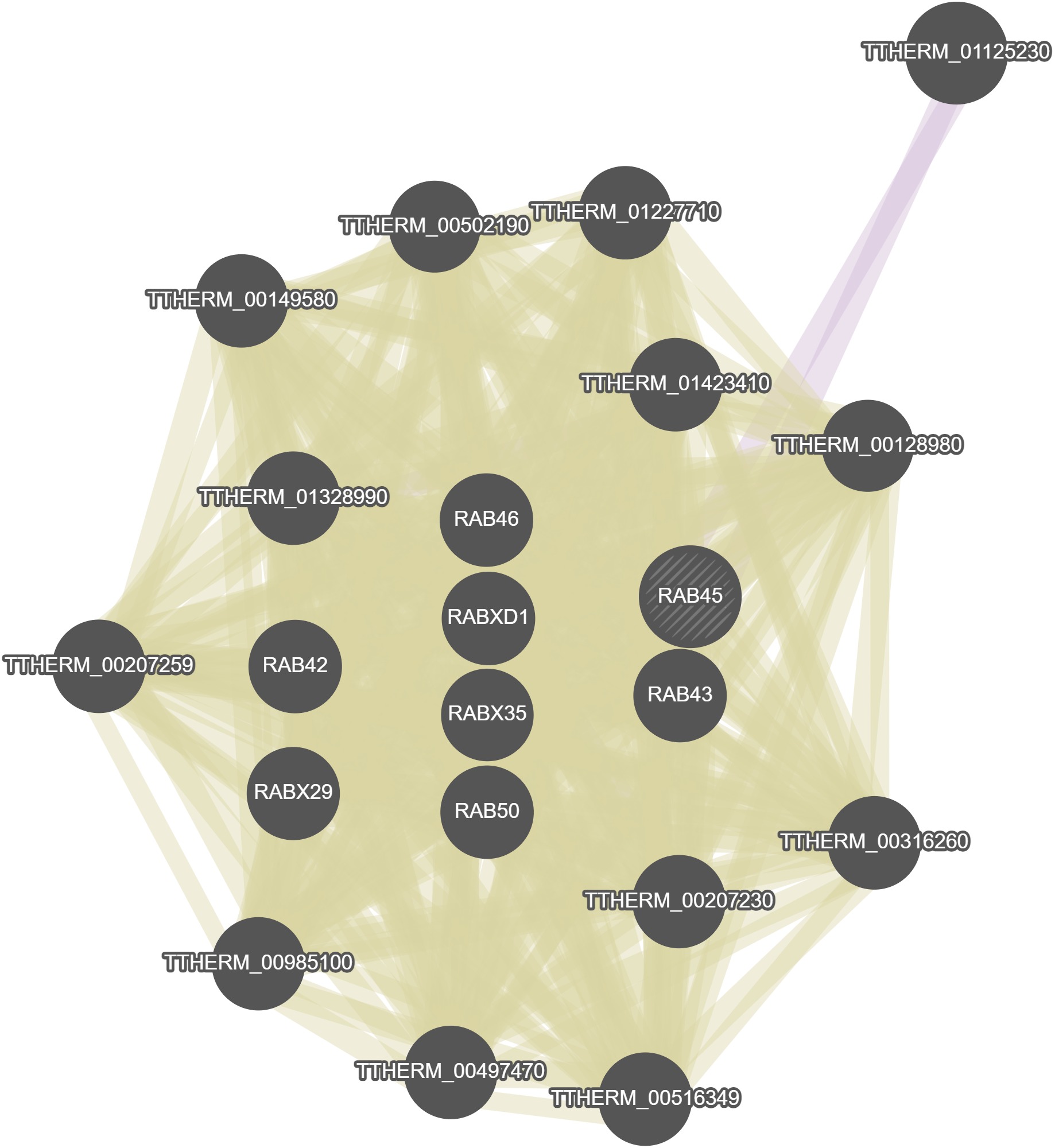
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:16933976: Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA,
 Sequences
Sequences
>TTHERM_00207260(coding)
ATGGATAAACTGCTTAATCAATCTAATTAAATAAAAATATTACTTTTCGGAGATTATTTG
ACAGGAAAATCTTCCTTGCTAATAAGATACTTTTAGGATAAGTTCTATGACTATCCTGAT
TTATTTAGGAGAGAATTTTACTAAAAAATTTTGAAAAGCGGAAAAGATGAAATTAATTTA
TACATATGTGAATTTGATTCAAATGAAAGAATGTACTACATAAAATAATTTTTTTATAGG
ACTTCTTCATTTATAATAATTTGTTATGATGTTACTAATAGATAAAGTTTTTAAAAAGCA
TATAATTGGTACTTAGATTTTAAGAATAGCAGCAATAATAAATATGTAGTTTTATCTTTA
ATTGGAAATAAATGTGATTTGCAGGAAGAACATAGGTAGGTTTCTTATCAAGAGGCTTAA
CAGCTTGCTTAATCCTTGGGAATGGTCTTCTTTGAAATTTCAGCTAAAACTGGTTATCAT
GTAAACGAGTTGTTTGATTTTATAATTACTTAGGCTCAAAATATTGCTTGA>TTHERM_00207260(gene)
AAAGCTTAATTAAATAATTTTTTTTCAATTTTTAATTAAGAAGTAATACCACAATAAATG
GATAAACTGCTTAATCAATCTAATTAAATAAAAATATTACTTTTCGGAGATTATTTGACA
GGAAAATCTTCCTTGCTAATAAGATACTTTTAGGATAAGTTCTATGACTATCCTGATTTA
TTTAGGAGAGAATTTTACTAAAAAATTTTGAAAAGCGGAAAAGATGAAATTAATTTATAC
ATATGTGAATTTGATTCAAATGAAAGAATGTACTACATAAAATAATTTTTTTATAGGACT
TCTTCATTTATAATAATTTGTTATGATGTTACTAATAGATAAAGTTTTTAAAAAGCATAT
AATTGGTACTTAGATTTTAAGAATAGCAGCAATAATAAATATGTAGTTTTATCTTTAATT
GGAAATAAATGTGATTTGCAGGAAGAACATAGGTAGGTTTCTTATCAAGAGGCTTAACAG
CTTGCTTAATCCTTGGGAATGGTCTTCTTTGAAATTTCAGCTAAAACTGGTTATCATGTA
AACGAGTTGTTTGATTTTATAATTACTTAGGCTCAAAATATTGCTTGAAAATGTCTAAAA
GCTAAATATTTTCTTAAGATAAAATGTATTAAATGTGGTTAAGTTAGGTATACTATTCAT
CTATATTTAAAAGCAACAAAAATCTAAAAACTATTATATTTTTAATCTAAATAATTGAGT
AAATAATATTTTTAAAGTTAAATTATTAGTTCTACAAGGTTTTAATAGATTCTAACTAAA
ACCTTTTTAGATATTTCAACTATCAAATTCCTGAATAAATATCTAATTAATTCCAAAAAA
AAAAAATGTGTGCAATTGTTAATAGTAAATAAAGAAATTAAATTTTTAAAATAAATCCTT
TTAGCTAATTTAAATTAAAATATGAAATTTATTCAAATATTTTGTTAGATTTTTTGAAGA
TAAAATCAATTTATTTAAAAAT>TTHERM_00207260(protein)
MDKLLNQSNQIKILLFGDYLTGKSSLLIRYFQDKFYDYPDLFRREFYQKILKSGKDEINL
YICEFDSNERMYYIKQFFYRTSSFIIICYDVTNRQSFQKAYNWYLDFKNSSNNKYVVLSL
IGNKCDLQEEHRQVSYQEAQQLAQSLGMVFFEISAKTGYHVNELFDFIITQAQNIA