Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00245740Standard Name
NRK2 (NIMA-Related Kinase )Aliases
PreTt02469 | 21.m00300Description
NRK2 plant dual-specificity MAP kinase kinase family domain protein; NIMA (Never In Mitosis) related kinase; hypothetical proteinGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- acyl-CoA ceramide synthase complex (IEA) | GO:0061576
- aleurone grain membrane (IEA) | GO:0032578
- chloroplast ribosome (IEA) | GO:0043253
- CURI complex (IEA) | GO:0032545
- Dbf4-dependent protein kinase complex (IEA) | GO:0031431
- interleukin-4 receptor complex (IEA) | GO:0016516
- MICOS complex (IEA) | GO:0061617
- mitotic nuclear membrane microtubule tethering complex (IEA) | GO:0106084
- multivesicular body (IEA) | GO:0005771
- obsolete chromosome segregation-directing complex (IEA) | GO:0043913
- obsolete transforming growth factor beta type I receptor homodimeric complex (IEA) | GO:0070018
- obsolete transforming growth factor beta type II receptor homodimeric complex (IEA) | GO:0070019
- obsolete transforming growth factor beta1-type II receptor complex (IEA) | GO:0070020
- organellar chromatophore (IEA) | GO:0070111
- vacuolar membrane (IEA) | GO:0005774
Molecular Function
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-octaprenyl-6-methoxyphenol hydroxylase activity (IEA) | GO:0008681
- 2-oxoglutarate decarboxylase activity (IEA) | GO:0008683
- 2-oxopent-4-enoate hydratase activity (IEA) | GO:0008684
- 3-hydroxybutyryl-CoA dehydrogenase activity (IEA) | GO:0008691
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- ABC-type amino acid transporter activity (IEA) | GO:0015424
- ABC-type maltose transporter activity (IEA) | GO:0015423
- ABC-type oligosaccharide transporter activity (IEA) | GO:0015422
- antheraxanthin epoxidase activity (IEA) | GO:0052663
- aromatase activity (IEA) | GO:0008402
- arsenate reductase (glutaredoxin) activity (IEA) | GO:0008794
- axon guidance receptor activity (IEA) | GO:0008046
- C-5 sterol desaturase activity (IEA) | GO:0000248
- carboxymethylenebutenolidase activity (IEA) | GO:0008806
- Cys-tRNA(Pro) hydrolase activity (IEA) | GO:0043907
- G protein-coupled neurotensin receptor activity (IEA) | GO:0016492
- gastric inhibitory peptide receptor activity (IEA) | GO:0016519
- histone succinyltransferase activity (IEA) | GO:0106078
- isochorismate synthase activity (IEA) | GO:0008909
- K63-linked deubiquitinase activity (IEA) | GO:0061578
- kanamycin kinase activity (IEA) | GO:0008910
- L-aspartate oxidase activity (IEA) | GO:0008734
- obsolete D-lysine oxidase activity (IEA) | GO:0043912
- obsolete DNA topoisomerase IV activity (IEA) | GO:0008724
- obsolete protein tagging activity (IEA) | GO:0008638
- obsolete RNA polymerase I transcription regulator recruiting activity (IEA) | GO:0001186
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete tachykinin (IEA) | GO:0008648
- phosphatidylcholine:cardiolipin O-linoleoyltransferase activity (IEA) | GO:0032577
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- rhamnulose-1-phosphate aldolase activity (IEA) | GO:0008994
- Ser(Gly)-tRNA(Ala) hydrolase activity (IEA) | GO:0043908
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- sterol 14-demethylase activity (IEA) | GO:0008398
- telomerase RNA binding (IEA) | GO:0070034
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
Biological Process
- absorption of visible light (IEA) | GO:0016038
- calcium ion transmembrane transport via high voltage-gated calcium channel (IEA) | GO:0061577
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- ceramide phosphoethanolamine catabolic process (IEA) | GO:1905372
- ceramide phosphoethanolamine metabolic process (IEA) | GO:1905371
- corticotropin hormone secreting cell development (IEA) | GO:0060131
- cytosine transport (IEA) | GO:0015856
- energy quenching (IEA) | GO:1990066
- female courtship behavior (IEA) | GO:0008050
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- interleukin-28A secretion (IEA) | GO:0072628
- laminaritriose transport (IEA) | GO:2001097
- maltotetraose transport (IEA) | GO:2001099
- mesonephric juxtaglomerular apparatus development (IEA) | GO:0061212
- mesonephric smooth muscle tissue development (IEA) | GO:0061214
- mucopolysaccharide metabolic process (IEA) | GO:1903510
- negative regulation by organism of cell-mediated immune response of other organism involved in symbiotic interaction (IEA) | GO:0052278
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070108
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of thyroid hormone receptor activity (IEA) | GO:1904168
- Notch signaling pathway involved in negative regulation of venous endothelial cell fate commitment (IEA) | GO:2000796
- obsolete negative regulation of adhesion of symbiont to host epithelial cell (IEA) | GO:1905227
- obsolete negative regulation of response to extracellular stimulus (IEA) | GO:0032105
- obsolete positive regulation by symbiont of host phytoalexin production (IEA) | GO:0052344
- obsolete positive regulation of adhesion of symbiont to host epithelial cell (IEA) | GO:1905228
- obsolete positive regulation of nucleic acid-templated transcription (IEA) | GO:1903508
- obsolete positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response (IEA) | GO:0006990
- obsolete regulation of glucose-6-phosphatase activity (IEA) | GO:0032114
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- ommochrome biosynthetic process (IEA) | GO:0006727
- P granule disassembly (IEA) | GO:1903864
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070109
- positive regulation of peroxisome proliferator activated receptor signaling pathway (IEA) | GO:0035360
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of retrograde transport, endosome to Golgi (IEA) | GO:1905281
- positive regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903691
- priming of cellular response to stress (IEA) | GO:0080136
- prolactin secreting cell development (IEA) | GO:0060132
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070107
- regulation of L-arginine import into cell (IEA) | GO:1902826
- response to reactive oxygen species (IEA) | GO:0000302
- response to singlet oxygen (IEA) | GO:0000304
- seed dehydration (IEA) | GO:1990068
- somatotropin secreting cell development (IEA) | GO:0060133
- sterol metabolic process (IEA) | GO:0016125
- sterol regulatory element binding protein cleavage involved in ER-nuclear sterol response pathway (IEA) | GO:0006992
- sterol regulatory element binding protein import into nucleus involved in sterol depletion response (IEA) | GO:0006993
- stomach neuroendocrine cell differentiation (IEA) | GO:0061102
- terpenoid catabolic process (IEA) | GO:0016115
- tetrasaccharide transport (IEA) | GO:2001098
- vacuolar acidification (IEA) | GO:0007035
- xanthophyll biosynthetic process (IEA) | GO:0016123
- xanthophyll catabolic process (IEA) | GO:0016124
 Domains
Domains
No Data fetched for Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
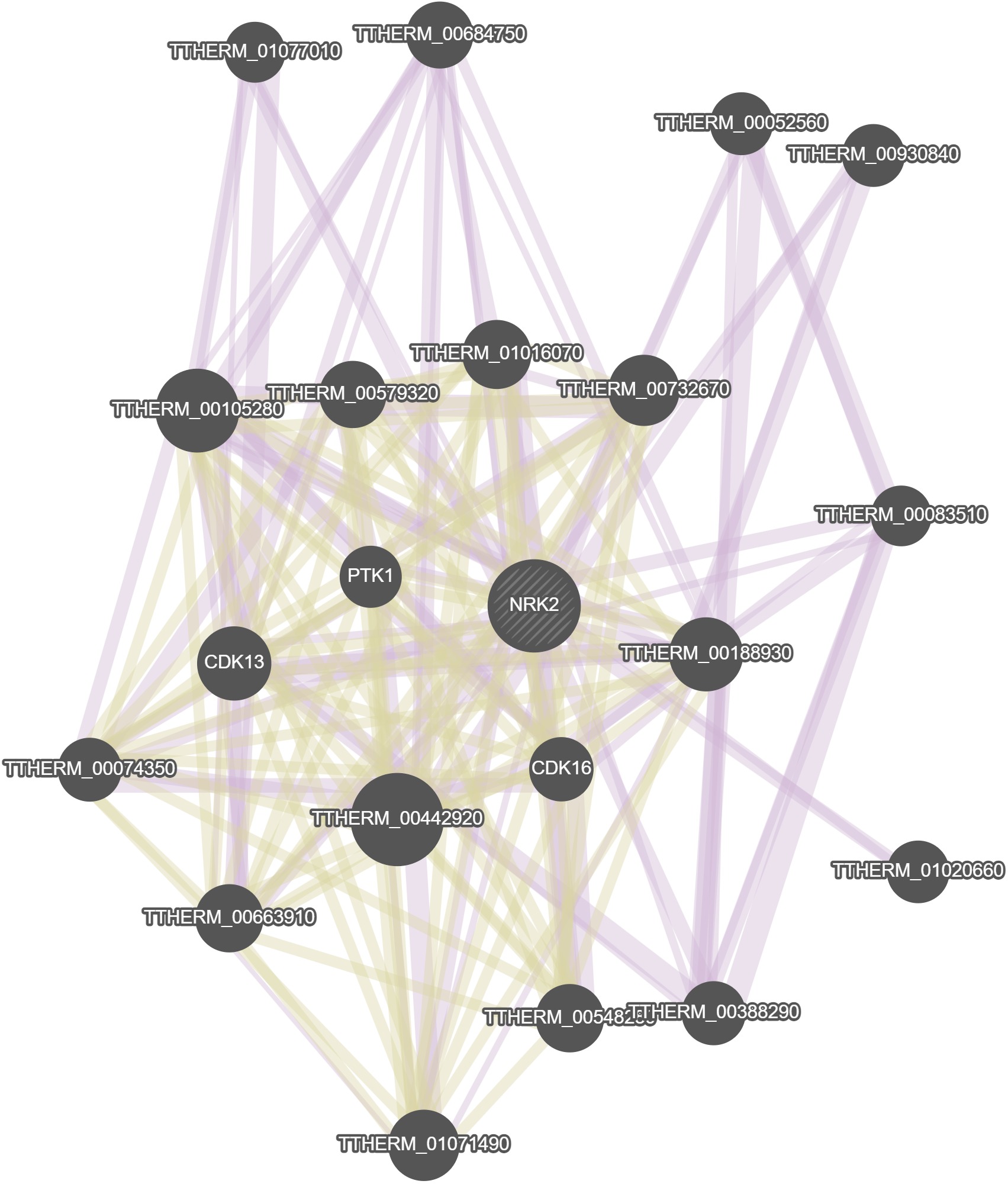
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:16611747: Wloga D, Camba A, Rogowski K, Manning G, Jerka-Dziadosz M, Gaertig J (2006) Members of the NIMA-related kinase family promote disassembly of cilia by multiple mechanisms. Molecular biology of the cell 17(6):2799-810
 Sequences
Sequences
>TTHERM_00245740(coding)
ATGTCCTTGAAAGATTTTGTGAATTTATAGAAGATAGGTGAGGGCTCTTACTCAAGTGTC
CATAAAGTAAGAAGGATCTCAGATAACTAAGAATACGCCCTCAAAAAGGTTAAATTATCT
GGTTTAAGCGAAAAAGAGAAAGATAACGCTCTTAATGAGATAAGAATATTAGCAAGTATA
GCTCATCCAAACATGATTGCATATAAGGATGCCTTTTTTGATGAATCTTCTCACTCACTT
TGCATTGTTATGGAGTTAGCCGTCAACGGTGATCTTTCTAAAAAAATTGATAGTGCAAAA
AAAAGAAATAGCTTTGTTCCAGAAGAAGAAATTTGGACAGTAGCGTTGCATATGTTAAGA
GGTTTGAAAGCTATGCACTCCAAAAAGATTCTTCACAGAGACCTCAAGTGTGCAAATGTC
TTTATTTCCAAGCAAGATGAATACAAAATTGGTGATCTCAATGTCTCAAAAGTCGCCAAA
AGAGGACTAGTATACACTTAAACTGGTACTCCTTATTACGCATCACCTGAAGTTTGGAGA
GATGAACCATATGACAGTTCAAGCGATATTTGGTCATTTGGATGCATTTTATACGAATTA
GCAGCTCTTAATCCACCATTTAGAGCTAAAGATATGGAAGGATTGTATAAAAAAGTATAG
AAAGGTATATTTGAGAGAATTCCATAAAGATATTCTAATGATTTGTAGAAGTTTATTGCA
TTATGCCTTTAGGTTAGTAGTGTATAACGTCCTTCTGTTACTTAGCTTTTAAACAATGTA
CTTCTTAAGCGTAATACCTAATTTTTAGCAGAATCTCCTGAAGAGTAATAAGATAAGGAA
AACACCCTTAGCCCAGAAGACCTTTTAAACACAATCAAAATGCCTAAAAACCCTCGTTAG
CTTAAGGAAAAGCTCCCAAAGCCTTAATATGATAAAGAGATATTAAATAATTCATATGAT
GGAGAAATACAACGCAAGGAGCGTAACCACAATAGGAATATGTCTGTAAAAATGAATAGG
ATATAATCTGTAACCAACCATAAATAGCATTAATACCTATAATAGTAATAGCAACTAATG
TTGGAGTAGGAAGGAAAAGATGTTGTTTTACCTTTAATTCCCAGATCATAATAATAAAAA
ATATTAGAACCTATAAGTAACAGCTAGTTACAGGCATAACAAAATAATAGCAATATTATA
GGTAGCTAAAATGATATTTATTCTAGAATAGGTAGCAACAGATATGAAAATTCTTCATCC
ATAGTTAGTGATTCCAGAAATTACTTGCAGGAAATACAGCCAAAAAAGCGAATTAATTAA
AATGGTTCAAGTGGTAACTACAATGAAGATTACTCCTAAAAATATAATAGATATGATTAA
CAACCAAGGTCAATAAACTCTCCAACTATATCTGTAATTTCTAAAAAGGATACAGGGTAT
TCAAATCCAGTGAACTCCATTAATCCACTAAGAAACAGAGTTAATTCTATAAATCAAATC
AATAGTTATGCAAGAATAGCTAATTTATATTAAAATTCTAAAAATCCCTATAACAACTAC
AGCGGAAATTCGATGCAGAGTGTTTCAATCGACTACAATCCTTAATATTCTCGTCCTGTT
TGGTGGGGTTGA>TTHERM_00245740(gene)
ATTGATAATTTCAATAATAAATACATAAGTTTATAAAAAGATAATCTAAGGATAATCAAT
ATTAATAACTAAGAGAGGATCAATCATTAATTTCTTTATAGTTTAATAGCAATAAACATA
TTTGAAGCAACAAAAAAACTAAAAAAGCTAACACCTGAATCATATTTAAATATTAATTTA
AAGCAAGTAATATAAGTAAATAAATGTCCTTGAAAGATTTTGTGAATTTATAGAAGATAG
GTGAGGGCTCTTACTCAAGTGTCCATAAAGTAAGAAGGATCTCAGATAACTAAGAATACG
CCCTCAAAAAGGTTAAATTATCTGGTTTAAGCGAAAAAGAGAAAGATAACGCTCTTAATG
AGATAAGAATATTAGCAAGTATAGCTCATCCAAACATGATTGCATATAAGGATGCCTTTT
TTGATGAATCTTCTCACTCACTTTGCATTGTTATGGAGTTAGCCGTCAACGGTGATCTTT
CTAAAAAAATTGATAGTGCAAAAAAAAGAAATAGCTTTGTTCCAGAAGAAGAAATTTGGA
CAGTAGCGTTGCATATGTTAAGAGGTTTGAAAGCTATGCACTCCAAAAAGATTCTTCACA
GAGACCTCAAGTGTGCAAATGTCTTTATTTCCAAGCAAGATGAATACAAAATTGGTGATC
TCAATGTCTCAAAAGTCGCCAAAAGAGGACTAGTATACACTTAAACTGGTACTCCTTATT
ACGCATCACCTGAAGTTTGGAGAGATGAACCATATGACAGTTCAAGCGATATTTGGTCAT
TTGGATGCATTTTATACGAATTAGCAGCTCTTAATCCACCATTTAGAGCTAAAGATATGG
AAGGATTGTATAAAAAAGTATAGAAAGGTATATTTGAGAGAATTCCATAAAGATATTCTA
ATGATTTGTAGAAGTTTATTGCATTATGCCTTTAGGTTAGTAGTGTATAACGTCCTTCTG
TTACTTAGCTTTTAAACAATGTACTTCTTAAGCGTAATACCTAATTTTTAGCAGAATCTC
CTGAAGAGTAATAAGATAAGGAAAACACCCTTAGCCCAGAAGACCTTTTAAACACAATCA
AAATGCCTAAAAACCCTCGTTAGCTTAAGGAAAAGCTCCCAAAGCCTTAATATGATAAAG
AGATATTAAATAATTCATATGATGGAGAAATACAACGCAAGGAGCGTAACCACAATAGGA
ATATGTCTGTAAAAATGAATAGGATATAATCTGTAACCAACCATAAATAGCATTAATACC
TATAATAGTAATAGCAACTAATGTTGGAGTAGGAAGGAAAAGATGTTGTTTTACCTTTAA
TTCCCAGATCATAATAATAAAAAATATTAGAACCTATAAGTAACAGCTAGTTACAGGCAT
AACAAAATAATAGCAATATTATAGGTAGCTAAAATGATATTTATTCTAGAATAGGTAGCA
ACAGATATGAAAATTCTTCATCCATAGTTAGTGATTCCAGAAATTACTTGCAGGAAATAC
AGCCAAAAAAGCGAATTAATTAAAATGGTTCAAGTGGTAACTACAATGAAGATTACTCCT
AAAAATATAATAGATATGATTAACAACCAAGGTCAATAAACTCTCCAACTATATCTGTAA
TTTCTAAAAAGGATACAGGGTATTCAAATCCAGTGAACTCCATTAATCCACTAAGAAACA
GAGTTAATTCTATAAATCAAATCAATAGTTATGCAAGAATAGCTAATTTATATTAAAATT
CTAAAAATCCCTATAACAACTACAGCGGAAATTCGATGCAGAGTGTTTCAATCGACTACA
ATCCTTAATATTCTCGTCCTGTTTGGTGGGGTTGATTGTATTCTTTTCTTGTTTTTATCA
AAGACATTTTGAGTTTATTTTTAAGATAGAAACAATAATAATTAATATGGATGACCAATA
AATAATAATTAAAATAATATTCAAAAAAACAATAATTAATCTAAATCACTCAAAAATAAT
AATTAATATACTACAAAAAATATTTTACAAATAAATAATAATTTCTTCTCTAATCATTCA
TTCATTAATATCTTCATAATTAAATTAAATATTAATTAGTTAATATCTTCTTCTCCTTCA
AATATTGTTTATAATTTGAATCTTCAAATGAAGATAC>TTHERM_00245740(protein)
MSLKDFVNLQKIGEGSYSSVHKVRRISDNQEYALKKVKLSGLSEKEKDNALNEIRILASI
AHPNMIAYKDAFFDESSHSLCIVMELAVNGDLSKKIDSAKKRNSFVPEEEIWTVALHMLR
GLKAMHSKKILHRDLKCANVFISKQDEYKIGDLNVSKVAKRGLVYTQTGTPYYASPEVWR
DEPYDSSSDIWSFGCILYELAALNPPFRAKDMEGLYKKVQKGIFERIPQRYSNDLQKFIA
LCLQVSSVQRPSVTQLLNNVLLKRNTQFLAESPEEQQDKENTLSPEDLLNTIKMPKNPRQ
LKEKLPKPQYDKEILNNSYDGEIQRKERNHNRNMSVKMNRIQSVTNHKQHQYLQQQQQLM
LEQEGKDVVLPLIPRSQQQKILEPISNSQLQAQQNNSNIIGSQNDIYSRIGSNRYENSSS
IVSDSRNYLQEIQPKKRINQNGSSGNYNEDYSQKYNRYDQQPRSINSPTISVISKKDTGY
SNPVNSINPLRNRVNSINQINSYARIANLYQNSKNPYNNYSGNSMQSVSIDYNPQYSRPV
WWG