Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00263330Standard Name
KIN2 (KINesin)Aliases
PreTt29398 | 23.m00294Description
KIN2 kinesin motor catalytic domain protein; Kinesin-II heavy chain homolog; implicated in intraciliary transport; shares essential functions with KIN1 in ciliary assembly and maintenance, rotokinesis and phagocytosis; Kinesin-like proteinGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- acyl-CoA ceramide synthase complex (IEA) | GO:0061576
- alpha6-beta4 integrin-Shc-Grb2 complex (IEA) | GO:0070333
- catalase complex (IEA) | GO:0062151
- chloroplast ribosome (IEA) | GO:0043253
- Dbf4-dependent protein kinase complex (IEA) | GO:0031431
- egg chorion (IEA) | GO:0042600
- inner plaque of mitotic spindle pole body (IEA) | GO:0061497
- kinesin II complex (ISS) | GO:0016939
- low-density lipoprotein receptor complex (IEA) | GO:0062136
- obsolete integral component of mitochondrial membrane (IEA) | GO:0032592
- outer plaque of mitotic spindle pole body (IEA) | GO:0061499
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-octaprenyl-6-methoxyphenol hydroxylase activity (IEA) | GO:0008681
- 2-oxoglutarate decarboxylase activity (IEA) | GO:0008683
- 2-oxopent-4-enoate hydratase activity (IEA) | GO:0008684
- 3-beta-hydroxy-delta5-steroid dehydrogenase (NAD+) activity (IEA) | GO:0003854
- 3-hydroxyisobutyryl-CoA hydrolase activity (IEA) | GO:0003860
- ABC-type ferric iron transporter activity (IEA) | GO:0015408
- ABC-type maltose transporter activity (IEA) | GO:0015423
- ABC-type monosaccharide transporter activity (IEA) | GO:0015407
- ABC-type oligosaccharide transporter activity (IEA) | GO:0015422
- aminotriazole transmembrane transporter activity (IEA) | GO:1901478
- antheraxanthin epoxidase activity (IEA) | GO:0052663
- arsenate reductase (glutaredoxin) activity (IEA) | GO:0008794
- benomyl transporter activity (IEA) | GO:0015242
- benzodiazepine receptor activity (IEA) | GO:0008503
- C-5 sterol desaturase activity (IEA) | GO:0000248
- carbamate kinase activity (IEA) | GO:0008804
- carboxymethylenebutenolidase activity (IEA) | GO:0008806
- carboxyvinyl-carboxyphosphonate phosphorylmutase activity (IEA) | GO:0008807
- catechol O-methyltransferase activity (IEA) | GO:0016206
- chemokine (C-C motif) ligand 12 binding (IEA) | GO:0035716
- copper-exporting ATPase activity (IEA) | GO:0004008
- corticosterone binding (IEA) | GO:1903875
- Cys-tRNA(Pro) hydrolase activity (IEA) | GO:0043907
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- ent-pimara-9(11),15-diene synthase activity (IEA) | GO:0052674
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- galactitol-1-phosphate 5-dehydrogenase activity (IEA) | GO:0008868
- glucosidase activity (IEA) | GO:0015926
- growth hormone-releasing hormone receptor activity (IEA) | GO:0016520
- insulin-like growth factor II binding (IEA) | GO:0031995
- interleukin-13 receptor activity (IEA) | GO:0016515
- inulin binding (IEA) | GO:2001082
- K63-linked deubiquitinase activity (IEA) | GO:0061578
- L-fucose isomerase activity (IEA) | GO:0008736
- L-fuculose-phosphate aldolase activity (IEA) | GO:0008738
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- lipopolysaccharide heptosyltransferase activity (IEA) | GO:0008920
- mercury ion transmembrane transporter activity (IEA) | GO:0015097
- methane monooxygenase [NAD(P)H] activity (IEA) | GO:0015049
- nonselective channel activity (IEA) | GO:0015249
- obsolete (3R)-3-hydroxybutyryl-CoA dehydratase activity (IEA) | GO:0003859
- obsolete amiloride transmembrane transporter activity (IEA) | GO:0015240
- obsolete aminoacetone:oxygen oxidoreductase(deaminating) activity (IEA) | GO:0052594
- obsolete aspartic-type signal peptidase activity (IEA) | GO:0009049
- obsolete D-lysine oxidase activity (IEA) | GO:0043912
- obsolete drug:proton antiporter activity (IEA) | GO:0015307
- obsolete protein tagging activity (IEA) | GO:0008638
- obsolete signal transducer, downstream of receptor, with protein tyrosine kinase activity (IEA) | GO:0004716
- obsolete sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity (IEA) | GO:0052591
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete tachykinin (IEA) | GO:0008648
- obsolete tocotrienol omega-hydroxylase activity (IEA) | GO:0052872
- obsolete tryptamine:oxygen oxidoreductase (deaminating) activity (IEA) | GO:0052593
- obsolete tubulin GTPase activity (IEA) | GO:0008550
- obsolete zinc, cadmium uptake permease activity (IEA) | GO:0015340
- oxidoreductase activity, acting on CH or CH2 groups, with an iron-sulfur protein as acceptor (IEA) | GO:0052592
- peptide antigen binding (IEA) | GO:0042605
- phosphatidylinositol-3-phosphate binding (IEA) | GO:0032266
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- pituitary adenylate cyclase activating polypeptide activity (IEA) | GO:0016521
- protein tyrosine kinase activity (IEA) | GO:0004713
- rRNA methyltransferase activity (IEA) | GO:0008649
- Ser(Gly)-tRNA(Ala) hydrolase activity (IEA) | GO:0043908
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- succinyldiaminopimelate transaminase activity (IEA) | GO:0009016
- thrombospondin receptor activity (IEA) | GO:0070053
- titin binding (IEA) | GO:0031432
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
- undecaprenol kinase activity (IEA) | GO:0009038
- zinc efflux active transmembrane transporter activity (IEA) | GO:0015341
Biological Process
- (R)-mevalonic acid biosynthetic process (IEA) | GO:1901737
- 10-formyltetrahydrofolate catabolic process (IEA) | GO:0009258
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- absorption of visible light (IEA) | GO:0016038
- actomyosin contractile ring actin filament bundle assembly (IEA) | GO:0071519
- actomyosin contractile ring assembly actin filament bundle convergence (IEA) | GO:0071520
- adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process (IEA) | GO:0086023
- anaerobic respiration (IEA) | GO:0009061
- angiogenesis involved in wound healing (IEA) | GO:0060055
- antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent (IEA) | GO:0002480
- aromatic amino acid family biosynthetic process (IEA) | GO:0009073
- aspartate family amino acid biosynthetic process (IEA) | GO:0009067
- cell budding (IEA) | GO:0007114
- cell-cell adhesion involved in sealing an epithelial fold (IEA) | GO:0060607
- cellular response to vanadate(3-) (IEA) | GO:1902439
- cilium assembly (IGI) | GO:0042384
- copper ion export (IEA) | GO:0060003
- cytoplasmic translational termination (IEA) | GO:0002184
- deoxyribonucleoside monophosphate catabolic process (IEA) | GO:0009159
- detection of stimulus involved in Dma1-dependent checkpoint (IEA) | GO:1902419
- detection of tumor cell (IEA) | GO:0002355
- dichotomous subdivision of terminal units involved in lung branching (IEA) | GO:0060448
- early endosome to recycling endosome transport (IEA) | GO:0061502
- eating behavior (IEA) | GO:0042755
- embryonic brain development (IEA) | GO:1990403
- endomitotic cell cycle (IEA) | GO:0007113
- ethylene biosynthetic process (IEA) | GO:0009693
- Factor XII activation (IEA) | GO:0002542
- fatty acid primary amide biosynthetic process (IEA) | GO:0062112
- fibroblast growth factor receptor signaling pathway (IEA) | GO:0008543
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- glycolytic process from glycerol (IEA) | GO:0061613
- head kidney structural organization (IEA) | GO:0072119
- helper T cell chemotaxis (IEA) | GO:0035704
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- homogentisate metabolic process (IEA) | GO:1901999
- hydrogen peroxide catabolic process (IEA) | GO:0042744
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- initiation of appressorium formation (IEA) | GO:0075025
- interleukin-19 production (IEA) | GO:0032622
- interleukin-2 production (IEA) | GO:0032623
- interleukin-4-dependent isotype switching to IgE isotypes (IEA) | GO:0035708
- intraciliary retrograde transport (IEA) | GO:0035721
- L-alanine catabolic process (IEA) | GO:0042853
- laminaritriose transport (IEA) | GO:2001097
- leukocyte chemotaxis involved in immune response (IEA) | GO:0002233
- macromolecule biosynthetic process (IEA) | GO:0009059
- maintenance of left sidedness (IEA) | GO:0061969
- mitochondrial membrane organization (IEA) | GO:0007006
- mitotic cytokinetic cell separation (IEA) | GO:1902409
- mitotic nuclear division (IEA) | GO:0007067
- mitotic spindle assembly checkpoint signaling (IEA) | GO:0007094
- modulation by organism of defense-related cell wall thickening in other organism involved in symbiotic interaction (IEA) | GO:0052300
- mononuclear cell proliferation (IEA) | GO:0032943
- mRNA splicing, via endonucleolytic cleavage and ligation (IEA) | GO:0070054
- NADH dehydrogenase complex assembly (IEA) | GO:0010257
- negative regulation by organism of cell-mediated immune response of other organism involved in symbiotic interaction (IEA) | GO:0052278
- negative regulation by organism of defense-related jasmonic acid-mediated signal transduction pathway in other organism involved in symbiotic interaction (IEA) | GO:0052267
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of ATF6-mediated unfolded protein response (IEA) | GO:1903892
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of establishment or maintenance of cell polarity regulating cell shape (IEA) | GO:2000770
- negative regulation of interleukin-1 production (IEA) | GO:0032692
- negative regulation of interleukin-10 secretion (IEA) | GO:2001180
- negative regulation of intrinsic apoptotic signaling pathway (IEA) | GO:2001243
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of metanephric comma-shaped body morphogenesis (IEA) | GO:2000007
- negative regulation of metanephric nephron tubule epithelial cell differentiation (IEA) | GO:0072308
- negative regulation of natural killer cell proliferation involved in immune response (IEA) | GO:0032821
- negative regulation of peripheral B cell deletion (IEA) | GO:0002909
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phospholipid translocation (IEA) | GO:0061093
- negative regulation of phytoalexin metabolic process (IEA) | GO:0052321
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of response to drug (IEA) | GO:2001024
- negative regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process (IEA) | GO:0062026
- negative regulation of steroid hormone biosynthetic process (IEA) | GO:0090032
- negative regulation of vasculogenesis (IEA) | GO:2001213
- neutral lipid metabolic process (IEA) | GO:0006638
- nipple development (IEA) | GO:0060618
- Notch signaling pathway involved in negative regulation of venous endothelial cell fate commitment (IEA) | GO:2000796
- obsolete cellular response to Dizocilpine (IEA) | GO:1905107
- obsolete cellular response to ouabain (IEA) | GO:1905105
- obsolete histone succinylation (IEA) | GO:0106077
- obsolete modulation by host of symbiont adenylate cyclase activity (IEA) | GO:0075075
- obsolete modulation of peptidase activity in other organism involved in symbiotic interaction (IEA) | GO:0052198
- obsolete movement of cell or subcellular component (IGI) | GO:0006928
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete negative regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity (IEA) | GO:1904218
- obsolete negative regulation of diuresis (IEA) | GO:0003077
- obsolete negative regulation of histone H3-K18 acetylation (IEA) | GO:1902030
- obsolete positive chemotaxis in environment of other organism involved in symbiotic interaction (IEA) | GO:0052221
- obsolete positive regulation by host of symbiont defense response (IEA) | GO:0052197
- obsolete positive regulation by organism of defense-related symbiont reactive oxygen species production (IEA) | GO:0052349
- obsolete positive regulation by symbiont of host phytoalexin production (IEA) | GO:0052344
- obsolete positive regulation of adhesion of symbiont to host epithelial cell (IEA) | GO:1905228
- obsolete positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response (IEA) | GO:0006990
- obsolete regulation of cAMP-mediated signaling in response to host (IEA) | GO:0075174
- obsolete regulation of cellular ketone metabolic process by regulation of transcription from RNA polymerase II promoter (IEA) | GO:0072364
- obsolete regulation of establishment of turgor in appressorium (IEA) | GO:0075040
- obsolete regulation of memory (IEA) | GO:1905248
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- ommochrome biosynthetic process (IEA) | GO:0006727
- organelle organization (IEA) | GO:0006996
- P granule disassembly (IEA) | GO:1903864
- PEP carboxykinase C4 photosynthesis (IEA) | GO:0009764
- peptidoglycan catabolic process (IEA) | GO:0009253
- phagocytosis (IGI) | GO:0006909
- phagocytosis (IEA) | GO:0006909
- phenanthrene catabolic process via trans-9(R),10(R)-dihydrodiolphenanthrene (IEA) | GO:0018956
- photosynthesis, light harvesting (IEA) | GO:0009765
- photosynthesis, light harvesting in photosystem I (IEA) | GO:0009768
- phylloquinone metabolic process (IEA) | GO:0042374
- plant-type cell wall modification (IEA) | GO:0009827
- polyamine acetylation (IEA) | GO:0032917
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of antifungal innate immune response (IEA) | GO:1905036
- positive regulation of deacetylase activity (IEA) | GO:0090045
- positive regulation of epithelial cell proliferation involved in wound healing (IEA) | GO:0060054
- positive regulation of establishment or maintenance of cell polarity regulating cell shape (IEA) | GO:2000771
- positive regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation (IEA) | GO:2000704
- positive regulation of flavonoid biosynthetic process (IEA) | GO:0009963
- positive regulation of forebrain neuron differentiation (IEA) | GO:2000979
- positive regulation of granzyme A production (IEA) | GO:2000513
- positive regulation of hindgut contraction (IEA) | GO:0060450
- positive regulation of I-kappaB phosphorylation (IEA) | GO:1903721
- positive regulation of interleukin-10 secretion (IEA) | GO:2001181
- positive regulation of interleukin-20 production (IEA) | GO:0032744
- positive regulation of metanephric ureteric bud development (IEA) | GO:2001076
- positive regulation of mitotic sister chromatid segregation (IEA) | GO:0062033
- positive regulation of neurofibrillary tangle assembly (IEA) | GO:1902998
- positive regulation of ovulation (IEA) | GO:0060279
- positive regulation of plant epidermal cell differentiation (IEA) | GO:1903890
- positive regulation of Rap GTPase activity (IEA) | GO:0032854
- positive regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process (IEA) | GO:0062027
- positive regulation of the force of heart contraction by epinephrine (IEA) | GO:0003059
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- prostate induction (IEA) | GO:0060514
- protein localization to old growing cell tip (IEA) | GO:1903858
- quinone cofactor metabolic process (IEA) | GO:0042375
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of atrial cardiac muscle cell membrane depolarization (IEA) | GO:0060371
- regulation of attachment of mitotic spindle microtubules to kinetochore (IEA) | GO:1902423
- regulation of capsule polysaccharide biosynthetic process (IEA) | GO:0062084
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of cellular respiration (IEA) | GO:0043457
- regulation of cellular response to hepatocyte growth factor stimulus (IEA) | GO:2001112
- regulation of chemokine (C-C motif) ligand 20 production (IEA) | GO:1903884
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of flavonoid biosynthetic process (IEA) | GO:0009962
- regulation of hematopoietic progenitor cell differentiation (IEA) | GO:1901532
- regulation of interleukin-17-mediated signaling pathway (IEA) | GO:1903881
- regulation of membrane lipid metabolic process (IEA) | GO:1905038
- regulation of pentose-phosphate shunt (IEA) | GO:0043456
- regulation of PERK-mediated unfolded protein response (IEA) | GO:1903897
- regulation of phosphatidylcholine biosynthetic process (IEA) | GO:2001245
- regulation of relaxation of cardiac muscle (IEA) | GO:1901897
- regulation of SCF-dependent proteasomal ubiquitin-dependent protein catabolic process (IEA) | GO:0062025
- regulation of striated muscle contraction (IEA) | GO:0006942
- regulation of systemic arterial blood pressure by local renal renin-angiotensin (IEA) | GO:0003086
- regulation of transcriptional start site selection at RNA polymerase II promoter (IEA) | GO:0001178
- response to curcumin (IEA) | GO:1904643
- response to iron(II) ion (IEA) | GO:0010040
- response to methamphetamine hydrochloride (IEA) | GO:1904313
- response to reactive oxygen species (IEA) | GO:0000302
- response to singlet oxygen (IEA) | GO:0000304
- response to sterol depletion (IEA) | GO:0006991
- response to UV-C (IEA) | GO:0010225
- ribonucleotide catabolic process (IEA) | GO:0009261
- ribonucleotide metabolic process (IEA) | GO:0009259
- salicylic acid biosynthetic process (IEA) | GO:0009697
- septum digestion after cytokinesis (IGI) | GO:0000920
- serine family amino acid biosynthetic process (IEA) | GO:0009070
- serotonin secretion, neurotransmission (IEA) | GO:0060096
- signal transduction involved in meiotic DNA replication checkpoint (IEA) | GO:0072440
- somatotropin secreting cell differentiation (IEA) | GO:0060126
- spindle assembly involved in male meiosis (IEA) | GO:0007053
- spindle assembly involved in male meiosis I (IEA) | GO:0007054
- spindle assembly involved in male meiosis II (IEA) | GO:0007055
- spindle organization (IEA) | GO:0007051
- sterol regulatory element binding protein cleavage involved in ER-nuclear sterol response pathway (IEA) | GO:0006992
- striated muscle contraction (IEA) | GO:0006941
- sulfide oxidation, using sulfide dehydrogenase (IEA) | GO:0070222
- T-helper 1 cell chemotaxis (IEA) | GO:0035706
- tetrasaccharide transport (IEA) | GO:2001098
- transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript (IEA) | GO:0001180
- vacuolar acidification (IEA) | GO:0007035
- vitamin D catabolic process (IEA) | GO:0042369
- water-soluble vitamin biosynthetic process (IEA) | GO:0042364
- xanthophyll biosynthetic process (IEA) | GO:0016123
- xanthophyll catabolic process (IEA) | GO:0016124
 Domains
Domains
- ( PF00225 ) Kinesin motor domain
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
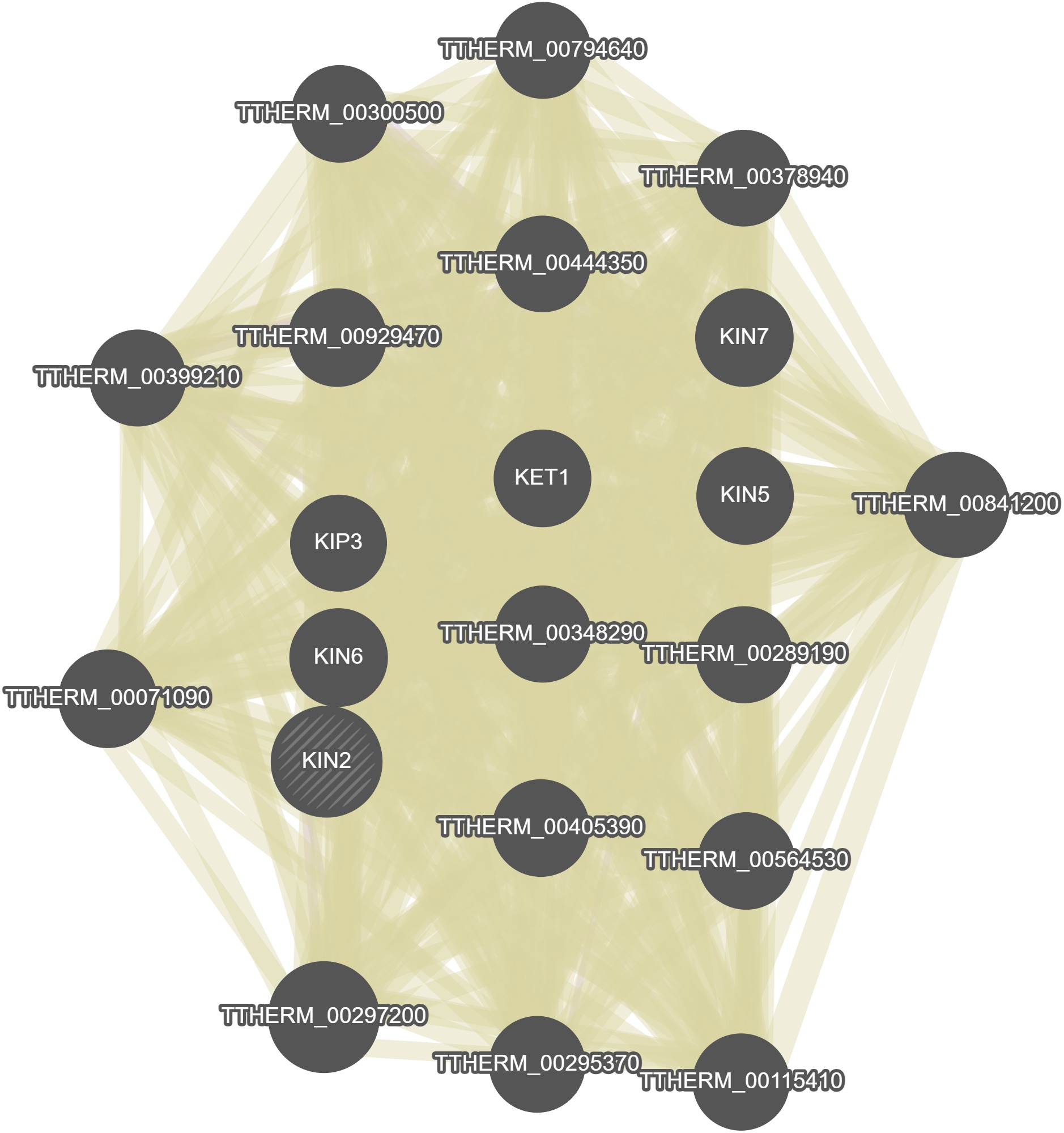
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:14983519: Awan A, Bernstein M, Hamasaki T, Satir P (2004) Cloning and characterization of Kin5, a novel Tetrahymena ciliary kinesin II. Cell motility and the cytoskeleton 58(1):1-9
- Ref:12925756: Brown JM, Fine NA, Pandiyan G, Thazhath R, Gaertig J (2003) Hypoxia regulates assembly of cilia in suppressors of Tetrahymena lacking an intraflagellar transport subunit gene. Molecular biology of the cell 14(8):3192-207
- Ref:10512852: Brown JM, Marsala C, Kosoy R, Gaertig J (1999) Kinesin-II is preferentially targeted to assembling cilia and is required for ciliogenesis and normal cytokinesis in Tetrahymena. Molecular biology of the cell 10(10):3081-96
- Ref:11212349: Cole DG (1999) Kinesin-II, the heteromeric kinesin. Cell Mol Life Sci 56(3-4):217-26
- Ref:10772758: Brown JM, Hardin C, Gaertig J (1999) Rotokinesis, a novel phenomenon of cell locomotion-assisted cytokinesis in the ciliate Tetrahymena thermophila. Cell biology international 23(12):841-8
 Sequences
Sequences
>TTHERM_00263330(coding)
ATGACTACAAGTAAAAACAACAGCGAATGCTTTAAGGTAGCTGTAAGATGCAGACCCCTA
AATAATGATGAAATATAAAATGAAAGAAGTTCAGTTGTTTCTGTAAACCCTCAAAGAGGA
GAAATATAAATCAAACAAAAGCATGATTAAGCTGAACAACATAGGATTTTTGCATTTGAC
TAAGTTTTCGAACCTGATATTTAATAAGAGGTTGTGTATAAATGTATTGCCTACCCTATT
GTTGAGAGCGTTTTAGAAGGCTACAATGGAACAATTTTCGCTTATGGCTAAACTGGTACT
GGAAAAACGCATACTATTTAAGGGAGAAATGAGCCAATTAATGAAAGAGGTATCATTCCT
AGGGCTTTTGAACATATTTTCCACTCTATTAAAGGCTCGCCAAACACTTAATTTTTAGTA
CATGTTAGTTTTCTTGAATTATATAATGAAGAGATTTAAGACTTGCTTTCCACCAAGAAT
AAAAAATTAGAATTAAGAGAAAAAGCAGAGACTGGCGTCTTTGTGAAAGATTTAACTAGT
TTTTTAGTTTAAAATGAATAGGAATTAAATGATAAATTTTAGCAAGGTATTTTGAATCGT
AAAGTTGGTTAAACTAAAATGAGTTCATGTTCATCTCGTTCTCACTCTATCCTTTCTGTA
ACTATTGAGAGGTGCGATGTTGTGAATGGGGAAAACCATATTAAAGTAGGTAAGTTAAAT
TTAGTTGACCTCGCTGGATCTGAACGTTAAAGTAAAACTCAAGCAACAGGCTCTCGTTTT
AAAGAAGGAGTATATATTAACTTAAGTTTAACTACTTTAGGTAATGTTATTTCCTCTCTC
ATCGATCCTAAAGCTTCTCATATTCCATATAGAGATTCTAAATTGACTAGAATTCTTTAA
GACTCTCTAGGTGGCAACACCAAAACTGTGATGATCGCTAATATTGGTCCTGCTGATTAT
AACTAAGATGAAACAATATCTACTTTAAGATATGCTCACAGAGCGAAATCAATATAAAAT
CATGCTTAAATTAACGAGGATCCCAAATAAGCAATGATAAGAAAATTTTAAGAGGAAATA
TCTAGCTTAAAATAGTAGTTATCAGGTTTACTTGAAACTGGAGGAGATTTTAATATGAAT
GCTGAAGTTAAAAAGATAGAAAAAATTGTTTTTAAATATGATGACGAAGAAATTATAAAA
TTGAAACAAAAACTTGAATAACAAAAATCTGATATTGAAAATAACTATGAAAATGAAGTA
AAGAAAATAGAAGAAGATAAGTAGTTACAGGAAAAAGAAAAGTAGCAGCTTATTTATTAG
CTTCAGGAAAAAGAACAAAAGTAGCAATAATAAAAATAGCAATAATAAAAACTACTTGGT
AAACTTTAGAAAATGTAACATAAAGTAATACAAGGAGAAGAAACTATGTAGAGAGTTCTT
GAAAACGAAAGATAACTGAATAAAGAATTGCAATAAGCAAATGAAAGAGATCGCTAAGCA
GCTTTACAAATCTAATAAAATGAAGATTTTATATTAGAGATAAACATGAAATTTAAGAAC
CAAGTTGAAGAAAAATAAGAAAAATAGAAAAGAATAAATCTTTTATTTGATAGATTGTGC
TAAATTTAATAATAAAATAAAGATATTGATGAATTTTATGTTTCTGAAATAGACTAGCTA
TAAGAAAGTTACAGAGACTTACAGAAAGAATTAAAATTAAAAAGATTTATCTTAGATTAT
TTCATTCCTTAAAAAGAATTAAAAAGATTATCAGAGAAAGCTATGTATAGTGAAGAATAA
CAAAATTGGGTATATCCTATTCAAAATTTGACTGGTAAATATATGAGAAGATCTGATGAA
AGTTAAGTTTCCCTTAAAGAAATTTAATCTGAAGATCTAGAACAAGATTCAAAAGTTTAT
TTTGTATACACAGAGCAAGGACCTATTAGAGAATAGTTTTTTTACACAACTGAAATAAAA
AAGCAAGAATATGCTTAACGAAAAAATGATCCTCTTAAACAAAATTTTGAATAATCAAAT
ACTTAATTTGTAAAAACTAAAAAGAGATCTAAAAGTTAAATAAATTAAATCTGA>TTHERM_00263330(gene)
AAAGAGATAAAATCATGACTACAAGTAAAAACAACAGCGAATGCTTTAAGGTAGCTGTAA
GATGCAGACCCCTAAATAATGATGAAATATAAAATGAAAGAAGTTCAGTTGTTTCTGTAA
ACCCTCAAAGAGGAGAAATATAAATCAAACAAAAGCATGATTAAGCTGAACAACATAGGA
TTTTTGCATTTGACTAAGTTTTCGAACCTGATATTTAATAAGAGGTTGTGTATAAATGTA
TTGCCTACCCTATTGTTGAGAGCGTTTTAGAAGGCTACAATGGAACAATTTTCGCTTATG
GCTAAACTGGTACTGGAAAAACGCATACTATTTAAGGGAGAAATGAGCCAATTAATGAAA
GAGGTATCATTCCTAGGGCTTTTGAACATATTTTCCACTCTATTAAAGGCTCGCCAAACA
CTTAATTTTTAGTACATGTTAGTTTTCTTGAATTATATAATGAAGAGATTTAAGACTTGC
TTTCCACCAAGAATAAAAAATTAGAATTAAGAGAAAAAGCAGAGACTGGCGTCTTTGTGA
AAGATTTAACTAGTTTTTTAGTTTAAAATGAATAGGAATTAAATGATAAATTTTAGCAAG
GTATTTTGAATCGTAAAGTTGGTTAAACTAAAATGAGTTCATGTTCATCTCGTTCTCACT
CTATCCTTTCTGTAACTATTGAGAGGTGCGATGTTGTGAATGGGGAAAACCATATTAAAG
TAGGTAAGTTAAATTTAGTTGACCTCGCTGGATCTGAACGTTAAAGTAAAACTCAAGCAA
CAGGCTCTCGTTTTAAAGAAGGAGTATATATTAACTTAAGTTTAACTACTTTAGGTAATG
TTATTTCCTCTCTCATCGATCCTAAAGCTTCTCATATTCCATATAGAGATTCTAAATTGA
CTAGAATTCTTTAAGACTCTCTAGGTGGCAACACCAAAACTGTGATGATCGCTAATATTG
GTCCTGCTGATTATAACTAAGATGAAACAATATCTACTTTAAGATATGCTCACAGAGCGA
AATCAATATAAAATCATGCTTAAATTAACGAGGATCCCAAATAAGCAATGATAAGAAAAT
TTTAAGAGGAAATATCTAGCTTAAAATAGTAGTTATCAGGTTTACTTGAAACTGGAGGAG
ATTTTAATATGAATGCTGAAGTTAAAAAGATAGAAAAAATTGTTTTTAAATATGATGACG
AAGAAATTATAAAATTGAAACAAAAACTTGAATAACAAAAATCTGATATTGAAAATAACT
ATGAAAATGAAGTAAAGAAAATAGAAGAAGATAAGTAGTTACAGGAAAAAGAAAAGTAGC
AGCTTATTTATTAGCTTCAGGAAAAAGAACAAAAGTAGCAATAATAAAAATAGCAATAAT
AAAAACTACTTGGTAAACTTTAGAAAATGTAACATAAAGTAATACAAGGAGAAGAAACTA
TGTAGAGAGTTCTTGAAAACGAAAGATAACTGAATAAAGAATTGCAATAAGCAAATGAAA
GAGATCGCTAAGCAGCTTTACAAATCTAATAAAATGAAGATTTTATATTAGAGATAAACA
TGAAATTTAAGAACCAAGTTGAAGAAAAATAAGAAAAATAGAAAAGAATAAATCTTTTAT
TTGATAGATTGTGCTAAATTTAATAATAAAATAAAGATATTGATGAATTTTATGTTTCTG
AAATAGACTAGCTATAAGAAAGTTACAGAGACTTACAGAAAGAATTAAAATTAAAAAGAT
TTATCTTAGATTATTTCATTCCTTAAAAAGAATTAAAAAGATTATCAGAGAAAGCTATGT
ATAGTGAAGAATAACAAAATTGGGTATATCCTATTCAAAATTTGACTGGTAAATATATGA
GAAGATCTGATGAAAGTTAAGTTTCCCTTAAAGAAATTTAATCTGAAGATCTAGAACAAG
ATTCAAAAGTTTATTTTGTATACACAGAGCAAGGACCTATTAGAGAATAGTTTTTTTACA
CAACTGAAATAAAAAAGCAAGAATATGCTTAACGAAAAAATGATCCTCTTAAACAAAATT
TTGAATAATCAAATACTTAATTTGTAAAAACTAAAAAGAGATCTAAAAGTTAAATAAATT
AAATCTGACTAAAAATAATTTTTTTAAATATCTAGTTATAATTTATTTTAAAAAAACTAT
TTATTAAGTTTAATAAAAAAATCACTTATTTGATATTGACAAAAAATATATTT>TTHERM_00263330(protein)
MTTSKNNSECFKVAVRCRPLNNDEIQNERSSVVSVNPQRGEIQIKQKHDQAEQHRIFAFD
QVFEPDIQQEVVYKCIAYPIVESVLEGYNGTIFAYGQTGTGKTHTIQGRNEPINERGIIP
RAFEHIFHSIKGSPNTQFLVHVSFLELYNEEIQDLLSTKNKKLELREKAETGVFVKDLTS
FLVQNEQELNDKFQQGILNRKVGQTKMSSCSSRSHSILSVTIERCDVVNGENHIKVGKLN
LVDLAGSERQSKTQATGSRFKEGVYINLSLTTLGNVISSLIDPKASHIPYRDSKLTRILQ
DSLGGNTKTVMIANIGPADYNQDETISTLRYAHRAKSIQNHAQINEDPKQAMIRKFQEEI
SSLKQQLSGLLETGGDFNMNAEVKKIEKIVFKYDDEEIIKLKQKLEQQKSDIENNYENEV
KKIEEDKQLQEKEKQQLIYQLQEKEQKQQQQKQQQQKLLGKLQKMQHKVIQGEETMQRVL
ENERQLNKELQQANERDRQAALQIQQNEDFILEINMKFKNQVEEKQEKQKRINLLFDRLC
QIQQQNKDIDEFYVSEIDQLQESYRDLQKELKLKRFILDYFIPQKELKRLSEKAMYSEEQ
QNWVYPIQNLTGKYMRRSDESQVSLKEIQSEDLEQDSKVYFVYTEQGPIREQFFYTTEIK
KQEYAQRKNDPLKQNFEQSNTQFVKTKKRSKSQINQI