Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00277530Standard Name
CDT1 (CDT1-like 1)Aliases
PreTt04794 | 25.m00258 | 3694.m00045Description
CDT1 DNA replication factor Cdt1; DNA replication factor CDT1; functions as part of DNA replication licensing complex; CDT1 Geminin-binding domain-likeGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- cellulosome (IEA) | GO:0043263
- chloroplast ribosome (IEA) | GO:0043253
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- macronucleus (IDA) | GO:0031039
- obsolete ectoplasm (IEA) | GO:0043265
- organellar chromatophore (IEA) | GO:0070111
- prospore membrane spindle pole body attachment site (IEA) | GO:0070057
- subrhabdomeral cisterna (IEA) | GO:0016029
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-octaprenyl-6-methoxyphenol hydroxylase activity (IEA) | GO:0008681
- 3-hydroxybutyrate dehydrogenase activity (IEA) | GO:0003858
- 3-hydroxybutyryl-CoA dehydrogenase activity (IEA) | GO:0008691
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- C-5 sterol desaturase activity (IEA) | GO:0000248
- carotene beta-ring hydroxylase activity (IEA) | GO:0010291
- CDP-diacylglycerol diphosphatase activity (IEA) | GO:0008715
- cell adhesive protein binding involved in atrial cardiac muscle cell-AV node cell communication (IEA) | GO:0086081
- cell adhesive protein binding involved in AV node cell-bundle of His cell communication (IEA) | GO:0086082
- delta-type opioid receptor binding (IEA) | GO:0031850
- dolichyl pyrophosphate Glc2Man9GlcNAc2 alpha-1,2-glucosyltransferase activity (IEA) | GO:0106073
- ferredoxin-NAD(P) reductase activity (IEA) | GO:0008937
- ferric triacetylfusarinine C:proton symporter activity (IEA) | GO:0015346
- FMN reductase (NADH) activity (IEA) | GO:0052874
- G protein-coupled neurotensin receptor activity (IEA) | GO:0016492
- JUN kinase phosphatase activity (IEA) | GO:0008579
- L-fucose isomerase activity (IEA) | GO:0008736
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- L-lysine transmembrane transporter activity (IEA) | GO:0015189
- lanosterol synthase activity (IEA) | GO:0000250
- lipopolysaccharide heptosyltransferase activity (IEA) | GO:0008920
- lipopolysaccharide-1,6-galactosyltransferase activity (IEA) | GO:0008921
- long-chain-3-hydroxyacyl-CoA dehydrogenase activity (IEA) | GO:0016509
- low voltage-gated potassium channel auxiliary protein activity (IEA) | GO:0015458
- mannitol-1-phosphate 5-dehydrogenase activity (IEA) | GO:0008926
- mercury ion transmembrane transporter activity (IEA) | GO:0015097
- metarhodopsin binding (IEA) | GO:0016030
- methane monooxygenase [NAD(P)H] activity (IEA) | GO:0015049
- mitochondrial single subunit type RNA polymerase activity (IEA) | GO:0001065
- MutSbeta complex binding (IEA) | GO:0032408
- obsolete aspartic-type signal peptidase activity (IEA) | GO:0009049
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete DNA topoisomerase IV activity (IEA) | GO:0008724
- obsolete peroxisome-assembly ATPase activity (IEA) | GO:0008573
- obsolete protein tagging activity (IEA) | GO:0008638
- obsolete RNA polymerase I transcription regulator recruiting activity (IEA) | GO:0001186
- obsolete sodium-translocating F-type ATPase activity (IEA) | GO:0016468
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete tachykinin (IEA) | GO:0008648
- obsolete tocopherol omega-hydroxylase activity (IEA) | GO:0052870
- obsolete X-opioid receptor activity (IEA) | GO:0015051
- obsolete zinc, iron permease activity (IEA) | GO:0015342
- oligoxyloglucan reducing-end-specific cellobiohydrolase activity (IEA) | GO:0033945
- oxalyl-CoA decarboxylase activity (IEA) | GO:0008949
- palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity (IEA) | GO:0008951
- penicillin amidase activity (IEA) | GO:0008953
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase (ATP) activity (IEA) | GO:0004612
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- phosphoglycerate dehydrogenase activity (IEA) | GO:0004617
- rhamnulose-1-phosphate aldolase activity (IEA) | GO:0008994
- siderophore uptake transmembrane transporter activity (IEA) | GO:0015344
- TFIID-class transcription factor complex binding (IEA) | GO:0001094
- thrombospondin receptor activity (IEA) | GO:0070053
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
- very-low-density lipoprotein particle receptor binding (IEA) | GO:0070326
- zinc efflux active transmembrane transporter activity (IEA) | GO:0015341
Biological Process
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- absorption of visible light (IEA) | GO:0016038
- basement membrane organization (IEA) | GO:0071711
- bud dilation involved in lung branching (IEA) | GO:0060503
- cell migration in diencephalon (IEA) | GO:0061381
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to D-galactose (IEA) | GO:1905378
- cellular response to dopamine (IEA) | GO:1903351
- cellular response to phosphate starvation (IEA) | GO:0016036
- ceramide phosphoethanolamine catabolic process (IEA) | GO:1905372
- ceramide phosphoethanolamine metabolic process (IEA) | GO:1905371
- coenzyme A biosynthetic process (IEA) | GO:0015937
- epithelial cell proliferation involved in lung bud dilation (IEA) | GO:0060505
- epithelium development (IEA) | GO:0060429
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- germinal center formation (IEA) | GO:0002467
- glycolytic process from glycerol (IEA) | GO:0061613
- glycolytic process through fructose-6-phosphate (IEA) | GO:0061615
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- hexasaccharide transport (IEA) | GO:2001102
- histoblast morphogenesis (IEA) | GO:0007488
- hydrogen transport (IEA) | GO:0006818
- imaginal disc-derived female genitalia development (IEA) | GO:0007486
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- induction by virus of host reticulophagy (IEA) | GO:0140883
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- inferior colliculus development (IEA) | GO:0061379
- interleukin-30 production (IEA) | GO:0072633
- interleukin-31 production (IEA) | GO:0072635
- internal genitalia morphogenesis (IEA) | GO:0035260
- L-alanine catabolic process (IEA) | GO:0042853
- leg disc morphogenesis (IEA) | GO:0007478
- lipid homeostasis (IEA) | GO:0055088
- lung connective tissue development (IEA) | GO:0060427
- lung pattern specification process (IEA) | GO:0060432
- lung saccule development (IEA) | GO:0060430
- maltoheptaose catabolic process (IEA) | GO:2001123
- maltohexaose transport (IEA) | GO:2001103
- mitochondrial membrane organization (IEA) | GO:0007006
- mitotic anaphase (IEA) | GO:0000090
- mitotic metaphase (IEA) | GO:0000089
- mitotic S phase (IEA) | GO:0000084
- mucopolysaccharide metabolic process (IEA) | GO:1903510
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of antigen processing and presentation of lipid antigen via MHC class Ib (IEA) | GO:0002599
- negative regulation of axon guidance (IEA) | GO:1902668
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of chemokine (C-C motif) ligand 2 secretion (IEA) | GO:1904208
- negative regulation of chemokine (C-C motif) ligand 20 production (IEA) | GO:1903885
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of epidermal growth factor receptor signaling pathway involved in heart process (IEA) | GO:1905283
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of ferulate catabolic process (IEA) | GO:1901467
- negative regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070108
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of peripheral B cell deletion (IEA) | GO:0002909
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phytoalexin metabolic process (IEA) | GO:0052321
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process (IEA) | GO:0062026
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- neutral lipid metabolic process (IEA) | GO:0006638
- Notch signaling pathway involved in negative regulation of venous endothelial cell fate commitment (IEA) | GO:2000796
- obsolete actomyosin contractile ring disassembly (IEA) | GO:1902621
- obsolete aerotaxis on or near other organism involved in symbiotic interaction (IEA) | GO:0052238
- obsolete aging (IEA) | GO:0007568
- obsolete catabolism by organism of cell wall pectin in other organism involved in symbiotic interaction (IEA) | GO:0052341
- obsolete extracellular carbohydrate transport (IEA) | GO:0006859
- obsolete fertilization, exchange of chromosomal proteins (IEA) | GO:0035042
- obsolete MAPK cascade involved in ascospore formation (IEA) | GO:1903695
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete negative regulation of adhesion of symbiont to host epithelial cell (IEA) | GO:1905227
- obsolete negative regulation of cytochrome-c oxidase activity (IEA) | GO:1905376
- obsolete oxidative single-stranded DNA demethylation (IEA) | GO:0035552
- obsolete positive regulation of adhesion of symbiont to host epithelial cell (IEA) | GO:1905228
- obsolete positive regulation of nucleic acid-templated transcription (IEA) | GO:1903508
- obsolete prothoracic morphogenesis (IEA) | GO:0007471
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete regulation of histone H3-K79 methylation (IEA) | GO:2001160
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete reproduction (IEA) | GO:0000003
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- obsolete sodium/potassium transport (IEA) | GO:0006815
- ommochrome biosynthetic process (IEA) | GO:0006727
- peripheral tolerance induction (IEA) | GO:0002465
- phagocytosis (IEA) | GO:0006909
- phosphate ion transport (IEA) | GO:0006817
- placenta blood vessel development (IEA) | GO:0060674
- plant-type cell wall modification (IEA) | GO:0009827
- poly(3-hydroxyalkanoate) biosynthetic process (IEA) | GO:0042621
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of antigen processing and presentation of peptide antigen via MHC class Ib (IEA) | GO:0002597
- positive regulation of dendrite extension (IEA) | GO:1903861
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of forebrain neuron differentiation (IEA) | GO:2000979
- positive regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070109
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of retrograde transport, endosome to Golgi (IEA) | GO:1905281
- positive regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process (IEA) | GO:0062027
- positive regulation of serine C-palmitoyltransferase activity (IEA) | GO:1904222
- positive regulation of skeletal muscle hypertrophy (IEA) | GO:1904206
- pronephric glomerulus morphogenesis (IEA) | GO:0035775
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- prothoracic disc morphogenesis (IEA) | GO:0007470
- quercetin biosynthetic process (IEA) | GO:1901734
- quercetin metabolic process (IEA) | GO:1901732
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of antigen processing and presentation of lipid antigen via MHC class Ib (IEA) | GO:0002598
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of hemicellulose catabolic process (IEA) | GO:2000988
- regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070107
- regulation of microglial cell migration (IEA) | GO:1904139
- regulation of phosphatidylcholine biosynthetic process (IEA) | GO:2001245
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of plant epidermal cell differentiation (IEA) | GO:1903888
- regulation of syringal lignin catabolic process (IEA) | GO:1901469
- regulation of transcription by RNA polymerase V (IEA) | GO:1904279
- regulation of urease activity (IEA) | GO:1905181
- release of sequestered calcium ion into cytosol by endoplasmic reticulum (IEA) | GO:1903514
- response to oxygen radical (IEA) | GO:0000305
- response to propan-1-ol (IEA) | GO:1901427
- response to reactive oxygen species (IEA) | GO:0000302
- response to singlet oxygen (IEA) | GO:0000304
- sodium-independent prostaglandin transport (IEA) | GO:0071720
- somatotropin secreting cell differentiation (IEA) | GO:0060126
- sperm aster formation (IEA) | GO:0035044
- sperm DNA decondensation (IEA) | GO:0035041
- striated muscle myosin thick filament assembly (IEA) | GO:0071688
- sulfur amino acid biosynthetic process (IEA) | GO:0000097
- sulfur amino acid metabolic process (IEA) | GO:0000096
- synaptic vesicle docking (IEA) | GO:0016081
- terpenoid catabolic process (IEA) | GO:0016115
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- trachea development (IEA) | GO:0060438
- transcription by RNA polymerase V (IEA) | GO:0001060
- triglyceride homeostasis (IEA) | GO:0070328
- wing disc morphogenesis (IEA) | GO:0007472
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
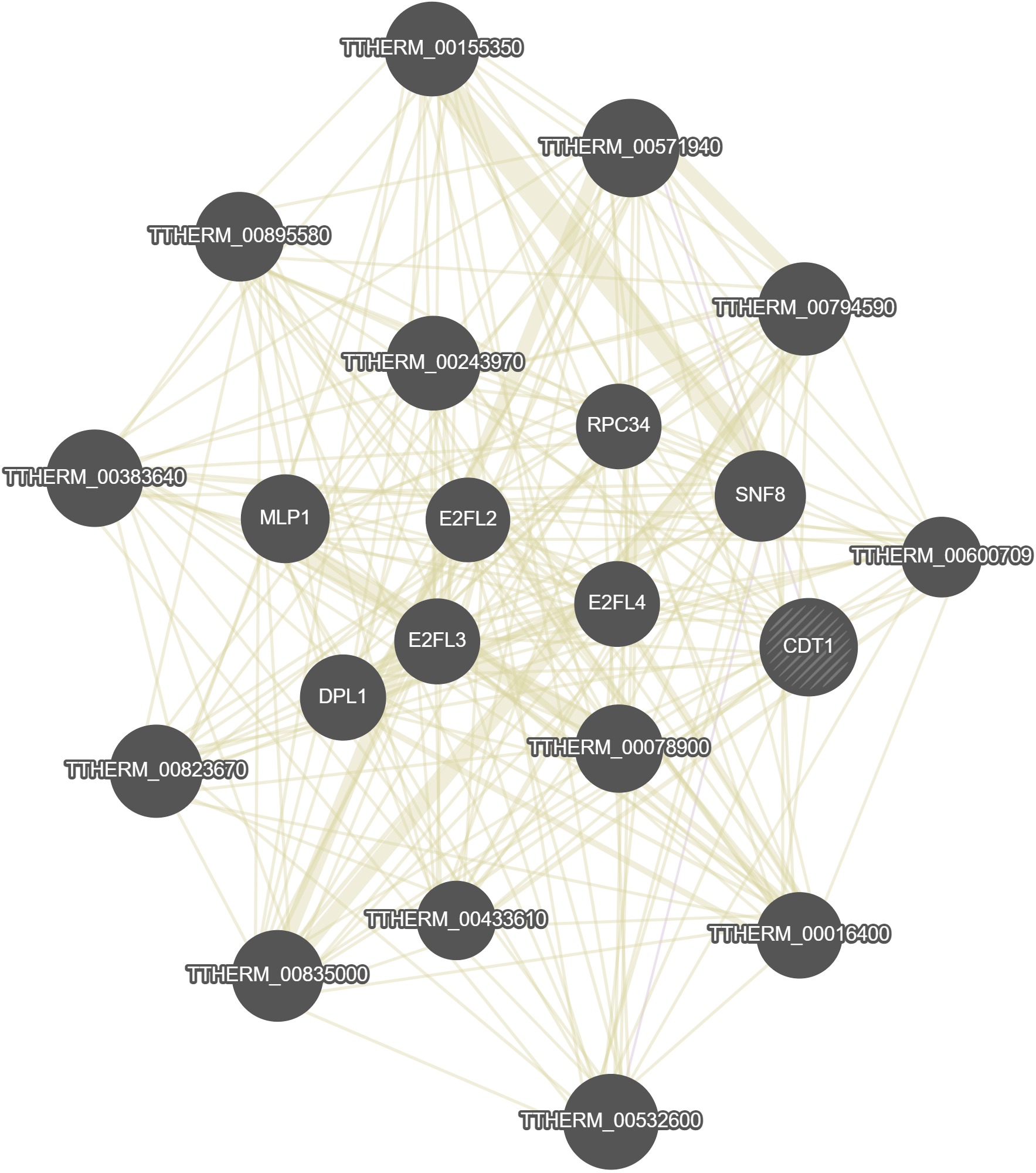
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00277530(coding)
ATGAGTTTAAGACAATAAACATTAACTCAATCCTTGGTTCCAGCTACTAATGAAGTAAGA
AAAGCTCCACAAACTAGAACAAGAGGAGGAGCTAAAAAAGAAATAGAGCTTGCTACTCCA
ATTACTGAATTTGTCAAAGGAAAGGAAACAAGAAAAAAAAAATTAAATGGTAAAAACTCT
AAAATTGATGATTATTATGATATCCATGGAAATGACAAAACCTTTATCTCGCATGATATT
GATAATGAAAAAGATCATAACCAATCAAGCATATCAACTGCCTCAACAAATACAACAATA
GATTATGAAAGAATTAATCATTTAAAGAGTATCAAATTAGCTTCTGATTTTAGTTAAATT
CCTTCTGGAAACACTATAACTATTGGAGGAAATTCTAATATTATGTAAGTCGAAGATAAG
CCTTTGCAAATAGATATTTAAGCAAATTAAAAACCTGCTATCGACTTTAAACAATTTTTA
GATGAGTAACAAAAGCAAAAACTCAAAAAAGAAGAAGAATTGGAGTAATAAAAATAACAA
CAGCAGTAAAAAAATGCTAAACTACTCTAAAATTTTGAGAATTTAAAAAAGAAATCTGCA
TTGTCTAATAGCACTACAGATTTAATATCAAAAATTTTAAACAAAAATAAAACTTAATAA
GACAAACCCGATAATATTTAAAATTTCAATAATCTTTCTTCTGCCAAAAAGATTGATTTG
ATTGAAGAAAAGCTTCTTTCAACACTTAAAATGAATTCTTCTGTTGAAAAGGATTCAAGC
AAAATAATGGATGCTGCTGATTAAATTTAAAAATTAAATCAAAAAGTTGAGCAATCCAAT
AAAATGATTGATGAAAAAGAAGAAATATTTGAACTTCAAACATAAAAGTTGAATTAAGTT
GATTCAGAAATTGAAATTACTAAATAAGAAGATAAAGCTGAAAGTAAATTTGAATAAATT
TTATAGACAGTTTAAAATAATGAAAAAGCTCAGAAAATTCTTGCTGGACCATCAATAAAG
CAAAGATATGAGGATTTATTGAAAGGTGATCAACAAGAATTTGCCTTACCCTACAAATAC
AAGGTGCTTTTAGAAACTTTTTCAAAGTTTGATGAGTGTATATAATTCTTTAAAGATAGA
AACAAACCAACTTTATTCAAAGATCTTAAATTTAATCTGGAACAAACTATGCGTGCCACT
ATCTCCCTTAAACAGATTTAGTAAATTATTCACTTGTACCCCTAAGCTTACTAGTTAAAG
TGGGTCTAAAATAAATAAGCTCATGAATAATTGGATTTATTAGTTAATTCTGCCTGTGAT
GAAAACGATAACAGAGTTGTTTTAACTAAAGCAGATCGTTTATAAAAACTTAGAAATTTA
CTTGTTGAAAAGGTTAAATATTTCCATAACTTGTACCTCAAATCTCAAGCTAGAGAGCAT
TTAGAGCAAGAGTTGTTAGATAACTTTAGATGGCATGATGATTTCAAGTTATAAGATGTC
CCCAATTTAGAAGAATAGGCTCTACCATTGAAACCTTTCGAAATTAAAGTTGAACCTCTA
AAAGAAACTTTGGAAAAGAATTAAGTAAAAAACGATATATTGGCTCGCTTAATGAAATCA
AAGGCTGAAGAATAAGAAGAAAAAGAAAAACAAAGAAAAGAGCTTATTGAAAAGAGTCTT
TATATTTCCACAAAAGAAAATAAGACAGGCTTGAGTGATCGTCTTTACCAGCTTATCAAG
GCTAAAGAAGAAAAAATTAATGAGGAAAAACTTAGATAATCTTTATCTAAAGGAACTGAA
ATGCAGAGAAGAATTCCTGAAATCAAAAAAGTTGTTGAAATTATAATGGTTTATTACATC
TAACGTAAAGTATCAAACATGTTTTATGCCTCTATTGTTGATTATCTCACTTAATCAAGC
AACATCATCATATCTAAAAGTTCTGCAATAGAAATGATTGATTATCTCATATAAAACTGC
CCCTTCTGGATAACTCTCATCCCAAATAAATAAGGAAGCATTATTAGAATTGACAAGAGT
GTTGCCCTTTCTAAAATCCTGGCAGAAATTGAAAAAATTGAAAGCTGA>TTHERM_00277530(gene)
AAAACAAAGATTAATAAAACACTTCTTTATTAGTTTATTAAATTATACAAAATTTACAAA
GAAAAATTATAAATAATAAGGCACAACTTCGAATAATTTGGCGCAAACATTCTTATAACA
AAATAAAAATCCATTGAAATAAAGCCACATAATTAATTATTCAAAAAGAAAGAAAGAAGC
AAACCTCTTATTTATTAGATTCAGAAGAAAATAATCAATACAAAACTAATAATAGTAAGA
CAAAAGATGAGTTTAAGACAATAAACATTAACTCAATCCTTGGTTCCAGCTACTAATGAA
GTAAGAAAAGCTCCACAAACTAGAACAAGAGGAGGAGCTAAAAAAGAAATAGAGCTTGCT
ACTCCAATTACTGAATTTGTCAAAGGAAAGGAAACAAGAAAAAAAAAATTAAATGGTAAA
AACTCTAAAATTGATGATTATTATGATATCCATGGAAATGACAAAACCTTTATCTCGCAT
GATATTGATAATGAAAAAGATCATAACCAATCAAGCATATCAACTGCCTCAACAAATACA
ACAATAGATTATGAAAGAATTAATCATTTAAAGAGTATCAAATTAGCTTCTGATTTTAGT
TAAATTCCTTCTGGAAACACTATAACTATTGGAGGAAATTCTAATATTATGTAAGTCGAA
GATAAGCCTTTGCAAATAGATATTTAAGCAAATTAAAAACCTGCTATCGACTTTAAACAA
TTTTTAGATGAGTAACAAAAGCAAAAACTCAAAAAAGAAGAAGAATTGGAGTAATAAAAA
TAACAACAGCAGTAAAAAAATGCTAAACTACTCTAAAATTTTGAGAATTTAAAAAAGAAA
TCTGCATTGTCTAATAGCACTACAGATTTAATATCAAAAATTTTAAACAAAAATAAAACT
TAATAAGACAAACCCGATAATATTTAAAATTTCAATAATCTTTCTTCTGCCAAAAAGATT
GATTTGATTGAAGAAAAGCTTCTTTCAACACTTAAAATGAATTCTTCTGTTGAAAAGGAT
TCAAGCAAAATAATGGATGCTGCTGATTAAATTTAAAAATTAAATCAAAAAGTTGAGCAA
TCCAATAAAATGATTGATGAAAAAGAAGAAATATTTGAACTTCAAACATAAAAGTTGAAT
TAAGTTGATTCAGAAATTGAAATTACTAAATAAGAAGATAAAGCTGAAAGTAAATTTGAA
TAAATTTTATAGACAGTTTAAAATAATGAAAAAGCTCAGAAAATTCTTGCTGGACCATCA
ATAAAGCAAAGATATGAGGATTTATTGAAAGGTGATCAACAAGAATTTGCCTTACCCTAC
AAATACAAGGTGCTTTTAGAAACTTTTTCAAAGTTTGATGAGTGTATATAATTCTTTAAA
GATAGAAACAAACCAACTTTATTCAAAGATCTTAAATTTAATCTGGAACAAACTATGCGT
GCCACTATCTCCCTTAAACAGATTTAGTAAATTATTCACTTGTACCCCTAAGCTTACTAG
TTAAAGTGGGTCTAAAATAAATAAGCTCATGAATAATTGGATTTATTAGTTAATTCTGCC
TGTGATGAAAACGATAACAGAGTTGTTTTAACTAAAGCAGATCGTTTATAAAAACTTAGA
AATTTACTTGTTGAAAAGGTTAAATATTTCCATAACTTGTACCTCAAATCTCAAGCTAGA
GAGCATTTAGAGCAAGAGTTGTTAGATAACTTTAGATGGCATGATGATTTCAAGTTATAA
GATGTCCCCAATTTAGAAGAATAGGCTCTACCATTGAAACCTTTCGAAATTAAAGTTGAA
CCTCTAAAAGAAACTTTGGAAAAGAATTAAGTAAAAAACGATATATTGGCTCGCTTAATG
AAATCAAAGGCTGAAGAATAAGAAGAAAAAGAAAAACAAAGAAAAGAGCTTATTGAAAAG
AGTCTTTATATTTCCACAAAAGAAAATAAGACAGGCTTGAGTGATCGTCTTTACCAGCTT
ATCAAGGCTAAAGAAGAAAAAATTAATGAGGAAAAACTTAGATAATCTTTATCTAAAGGA
ACTGAAATGCAGAGAAGAATTCCTGAAATCAAAAAAGTTGTTGAAATTATAATGGTTTAT
TACATCTAACGTAAAGTATCAAACATGTTTTATGCCTCTATTGTTGATTATCTCACTTAA
TCAAGCAACATCATCATATCTAAAAGTTCTGCAATAGAAATGATTGATTATCTCATATAA
AACTGCCCCTTCTGGATAACTCTCATCCCAAATAAATAAGGAAGCATTATTAGAATTGAC
AAGAGTGTTGCCCTTTCTAAAATCCTGGCAGAAATTGAAAAAATTGAAAGCTGATTGTAT
AGATTATTAAATAACAACTTAAGGTTAGAATGGAAAATAAACAATCAAACAGACAAACAT
ATTATTGTTTTCAAATGATTTAAAATAATTAAAAATAACTAAACTGTAATATAAATAAAG
AAGAAGCTATTTAAAACTAACTCAAAAGTAAATAATAGTTTGTTTCTTGAAATTTTTGCT
TTTCAAAAAATAAATTAAATGTATAAAAATTATAATTAAAATATAAATTAATTTAATTGA
TAAAACCATGTTAATAAATTAAAAAATATATAAATAAATACTTGTTGTTTTCATTAAATA
AATGTCCACAAAATATAACTCTTAAAAAATTATTAATATGATTGAATCTCAAACATTTAA
TGTCTAATTT>TTHERM_00277530(protein)
MSLRQQTLTQSLVPATNEVRKAPQTRTRGGAKKEIELATPITEFVKGKETRKKKLNGKNS
KIDDYYDIHGNDKTFISHDIDNEKDHNQSSISTASTNTTIDYERINHLKSIKLASDFSQI
PSGNTITIGGNSNIMQVEDKPLQIDIQANQKPAIDFKQFLDEQQKQKLKKEEELEQQKQQ
QQQKNAKLLQNFENLKKKSALSNSTTDLISKILNKNKTQQDKPDNIQNFNNLSSAKKIDL
IEEKLLSTLKMNSSVEKDSSKIMDAADQIQKLNQKVEQSNKMIDEKEEIFELQTQKLNQV
DSEIEITKQEDKAESKFEQILQTVQNNEKAQKILAGPSIKQRYEDLLKGDQQEFALPYKY
KVLLETFSKFDECIQFFKDRNKPTLFKDLKFNLEQTMRATISLKQIQQIIHLYPQAYQLK
WVQNKQAHEQLDLLVNSACDENDNRVVLTKADRLQKLRNLLVEKVKYFHNLYLKSQAREH
LEQELLDNFRWHDDFKLQDVPNLEEQALPLKPFEIKVEPLKETLEKNQVKNDILARLMKS
KAEEQEEKEKQRKELIEKSLYISTKENKTGLSDRLYQLIKAKEEKINEEKLRQSLSKGTE
MQRRIPEIKKVVEIIMVYYIQRKVSNMFYASIVDYLTQSSNIIISKSSAIEMIDYLIQNC
PFWITLIPNKQGSIIRIDKSVALSKILAEIEKIES