Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00297140Standard Name
RPL23 (Ribosomal Protein of the Large subunit #23)Aliases
PreTt15725 | 27.m00385 | 3692.m00187 | RPL14Description
RPL23 60S ribosomal protein L23; Homolog of Yeast RPL23, Human RPL23, Bacteria RPL14; Ribosomal protein L14p/L23e containing protein; Large ribosomal subunit protein uL14Genome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 11beta-hydroxyprogesterone binding (IEA) | GO:1903879
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- CCR10 chemokine receptor binding (IEA) | GO:0031735
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- prenyltransferase activity (IEA) | GO:0004659
- protein histidine kinase activity (IEA) | GO:0004673
Biological Process
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- microtubule depolymerization (IEA) | GO:0007019
- mitochondrial membrane organization (IEA) | GO:0007006
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of telomere single strand break repair (IEA) | GO:1903824
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete bent DNA binding (IEA) | GO:0006326
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- pigment accumulation in response to UV light (IEA) | GO:0043478
- pigment accumulation in tissues in response to UV light (IEA) | GO:0043479
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- TIRAP-dependent toll-like receptor 4 signaling pathway (IEA) | GO:0035665
 Domains
Domains
- ( PF00238 ) Ribosomal protein L14p/L23e
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
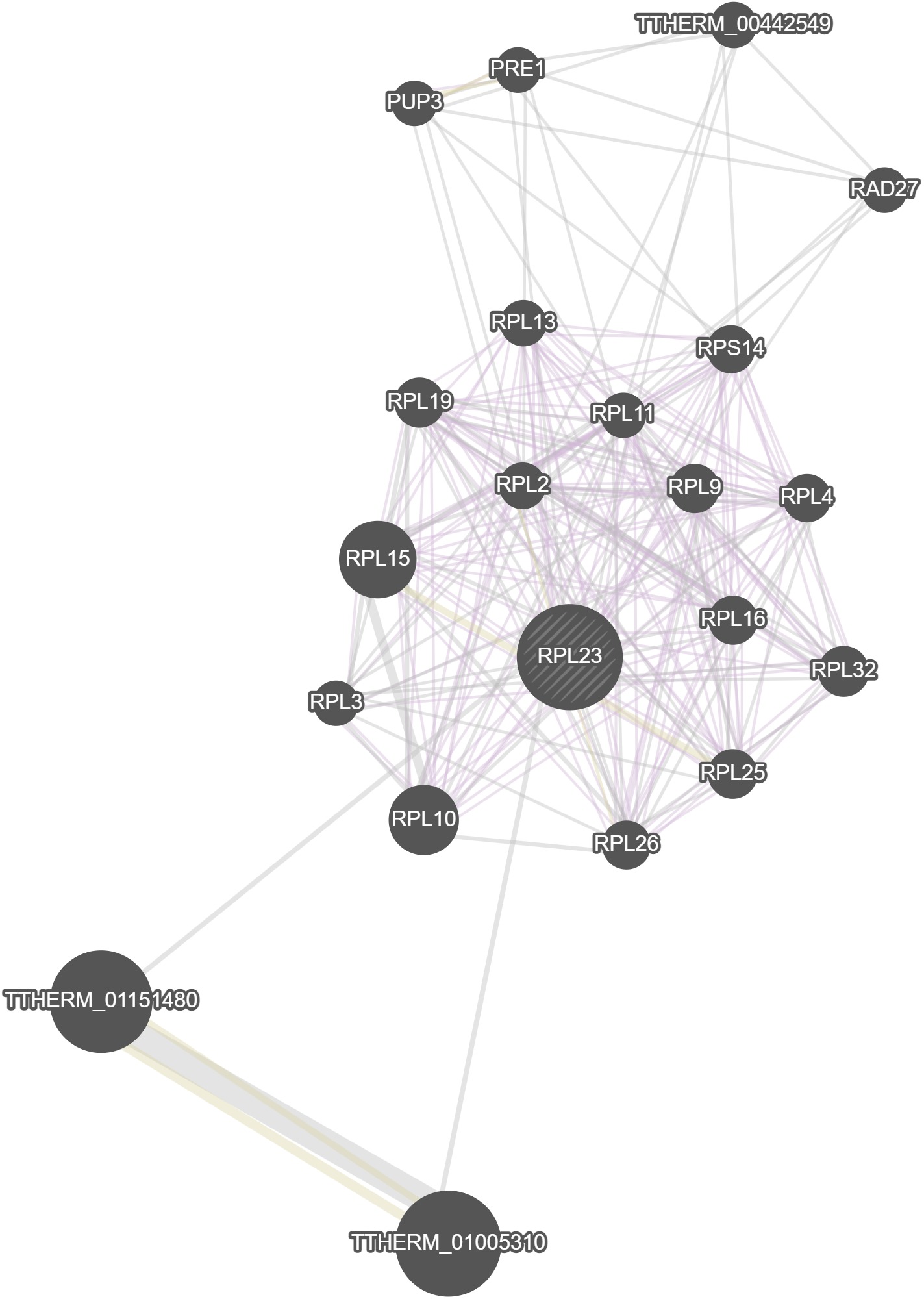
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:22052974: Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N (2011) Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science (New York, N.Y.) 334(6058):941-8
 Sequences
Sequences
>TTHERM_00297140(coding)
ATGGCTGCTGCTAGAGGTAGAGGTGGTTAAGTCGGTACCAAGGCTAAAGTTTCCTTGGGT
CTTCCCGTCGGTGCTGTTATGAACTGCGCTGATAACTCCGGTGCTAAGAACTTATATACT
ATTGCTTGTTTCGGTATCAAGGGTCACTTGAGCAAGCTCCCCAGTGCTTCCATTGGTGAT
ATGATTCTCTGCTCCGTCAAGAAGGGTTCCCCCAAGCTCAGAAAGAAAGTTCTCTAAGCT
ATCGTCATCAGACAAAGAAGACCCTGGAGAAGAAGAGATGGTGTCTTCATCTACTTCGAA
GATAATGCTGGTGTTATTGCTAACCCCAAGGGTGAAATGAAGGGTTCCCAAATCACTGGT
CCCGTCGCCAAGGAATGTGCTGACATCTGGCCCAAGGTTGCTTCCAACGCTGGTTCCGTT
GTTTGA>TTHERM_00297140(gene)
AAAGAAATACATAATACAAAATGGCTGCTGCTAGAGGTAGAGGTGGTTAAGTCGGTACCA
AGGCTAAAGTTTCCTTGGGTCTTCCCGTCGGTGCTGTTATGAACTGCGCTGATAACTCCG
GTGCTAAGAACTTATATACTATTGCTTGTTTCGGTATCAAGGGTCACTTGAGCAAGCTCC
CCAGTGCTTCCATTGGTGATATGATTCTCTGCTCCGTCAAGAAGGGTTCCCCCAAGCTCA
GAAAGAAAGTTCTCTAAGCTATCGTCATCAGACAAAGAAGACCCTGGAGAAGAAGAGATG
GTGTCTTCATCTACTTCGAAGATAATGCTGGTGTTATTGCTAACCCCAAGGGTGAAATGA
AGGGTTCCCAAATCACTGGTCCCGTCGCCAAGGAATGTGCTGACATCTGGCCCAAGGTTG
CTTCCAACGCTGGTTCCGTTGTTTGAAAAAAATAAAATATATAAAATTATAAACTTAAAT
GTATACTATTAAAATGTGAATACTTTTTTAAACTATAATTGCCAAAACTGTTATTTTACA
TATTTGTAGTATTAATATTTTTGGTTTTATTTAAATCAACC>TTHERM_00297140(protein)
MAAARGRGGQVGTKAKVSLGLPVGAVMNCADNSGAKNLYTIACFGIKGHLSKLPSASIGD
MILCSVKKGSPKLRKKVLQAIVIRQRRPWRRRDGVFIYFEDNAGVIANPKGEMKGSQITG
PVAKECADIWPKVASNAGSVV