Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00301840Standard Name
NRK5 (NIMA-Related Kinase )Aliases
PreTt19380 | 28.m00339 | 3691.m00156Description
NRK5 plant dual-specificity MAP kinase kinase family domain protein; NIMA-Related Kinase; NEK Ser/Thr protein kinase family, NIMA subfamilyGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- acyl-CoA ceramide synthase complex (IEA) | GO:0061576
- aleurone grain membrane (IEA) | GO:0032578
- chloroplast ribosome (IEA) | GO:0043253
- CURI complex (IEA) | GO:0032545
- Dbf4-dependent protein kinase complex (IEA) | GO:0031431
- interleukin-4 receptor complex (IEA) | GO:0016516
- MICOS complex (IEA) | GO:0061617
- mitotic nuclear membrane microtubule tethering complex (IEA) | GO:0106084
- multivesicular body (IEA) | GO:0005771
- obsolete chromosome segregation-directing complex (IEA) | GO:0043913
- obsolete transforming growth factor beta type I receptor homodimeric complex (IEA) | GO:0070018
- obsolete transforming growth factor beta type II receptor homodimeric complex (IEA) | GO:0070019
- obsolete transforming growth factor beta1-type II receptor complex (IEA) | GO:0070020
- organellar chromatophore (IEA) | GO:0070111
- vacuolar membrane (IEA) | GO:0005774
Molecular Function
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-octaprenyl-6-methoxyphenol hydroxylase activity (IEA) | GO:0008681
- 2-oxoglutarate decarboxylase activity (IEA) | GO:0008683
- 2-oxopent-4-enoate hydratase activity (IEA) | GO:0008684
- 3-hydroxybutyryl-CoA dehydrogenase activity (IEA) | GO:0008691
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- ABC-type amino acid transporter activity (IEA) | GO:0015424
- ABC-type maltose transporter activity (IEA) | GO:0015423
- ABC-type oligosaccharide transporter activity (IEA) | GO:0015422
- antheraxanthin epoxidase activity (IEA) | GO:0052663
- aromatase activity (IEA) | GO:0008402
- arsenate reductase (glutaredoxin) activity (IEA) | GO:0008794
- axon guidance receptor activity (IEA) | GO:0008046
- C-5 sterol desaturase activity (IEA) | GO:0000248
- carboxymethylenebutenolidase activity (IEA) | GO:0008806
- Cys-tRNA(Pro) hydrolase activity (IEA) | GO:0043907
- G protein-coupled neurotensin receptor activity (IEA) | GO:0016492
- gastric inhibitory peptide receptor activity (IEA) | GO:0016519
- histone succinyltransferase activity (IEA) | GO:0106078
- isochorismate synthase activity (IEA) | GO:0008909
- K63-linked deubiquitinase activity (IEA) | GO:0061578
- kanamycin kinase activity (IEA) | GO:0008910
- L-aspartate oxidase activity (IEA) | GO:0008734
- obsolete D-lysine oxidase activity (IEA) | GO:0043912
- obsolete DNA topoisomerase IV activity (IEA) | GO:0008724
- obsolete protein tagging activity (IEA) | GO:0008638
- obsolete RNA polymerase I transcription regulator recruiting activity (IEA) | GO:0001186
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete tachykinin (IEA) | GO:0008648
- phosphatidylcholine:cardiolipin O-linoleoyltransferase activity (IEA) | GO:0032577
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- rhamnulose-1-phosphate aldolase activity (IEA) | GO:0008994
- Ser(Gly)-tRNA(Ala) hydrolase activity (IEA) | GO:0043908
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- sterol 14-demethylase activity (IEA) | GO:0008398
- telomerase RNA binding (IEA) | GO:0070034
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
Biological Process
- absorption of visible light (IEA) | GO:0016038
- calcium ion transmembrane transport via high voltage-gated calcium channel (IEA) | GO:0061577
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- ceramide phosphoethanolamine catabolic process (IEA) | GO:1905372
- ceramide phosphoethanolamine metabolic process (IEA) | GO:1905371
- corticotropin hormone secreting cell development (IEA) | GO:0060131
- cytosine transport (IEA) | GO:0015856
- energy quenching (IEA) | GO:1990066
- female courtship behavior (IEA) | GO:0008050
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- interleukin-28A secretion (IEA) | GO:0072628
- laminaritriose transport (IEA) | GO:2001097
- maltotetraose transport (IEA) | GO:2001099
- mesonephric juxtaglomerular apparatus development (IEA) | GO:0061212
- mesonephric smooth muscle tissue development (IEA) | GO:0061214
- mucopolysaccharide metabolic process (IEA) | GO:1903510
- negative regulation by organism of cell-mediated immune response of other organism involved in symbiotic interaction (IEA) | GO:0052278
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070108
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of thyroid hormone receptor activity (IEA) | GO:1904168
- Notch signaling pathway involved in negative regulation of venous endothelial cell fate commitment (IEA) | GO:2000796
- obsolete negative regulation of adhesion of symbiont to host epithelial cell (IEA) | GO:1905227
- obsolete negative regulation of response to extracellular stimulus (IEA) | GO:0032105
- obsolete positive regulation by symbiont of host phytoalexin production (IEA) | GO:0052344
- obsolete positive regulation of adhesion of symbiont to host epithelial cell (IEA) | GO:1905228
- obsolete positive regulation of nucleic acid-templated transcription (IEA) | GO:1903508
- obsolete positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response (IEA) | GO:0006990
- obsolete regulation of glucose-6-phosphatase activity (IEA) | GO:0032114
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- ommochrome biosynthetic process (IEA) | GO:0006727
- P granule disassembly (IEA) | GO:1903864
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070109
- positive regulation of peroxisome proliferator activated receptor signaling pathway (IEA) | GO:0035360
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of retrograde transport, endosome to Golgi (IEA) | GO:1905281
- positive regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903691
- priming of cellular response to stress (IEA) | GO:0080136
- prolactin secreting cell development (IEA) | GO:0060132
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070107
- regulation of L-arginine import into cell (IEA) | GO:1902826
- response to reactive oxygen species (IEA) | GO:0000302
- response to singlet oxygen (IEA) | GO:0000304
- seed dehydration (IEA) | GO:1990068
- somatotropin secreting cell development (IEA) | GO:0060133
- sterol metabolic process (IEA) | GO:0016125
- sterol regulatory element binding protein cleavage involved in ER-nuclear sterol response pathway (IEA) | GO:0006992
- sterol regulatory element binding protein import into nucleus involved in sterol depletion response (IEA) | GO:0006993
- stomach neuroendocrine cell differentiation (IEA) | GO:0061102
- terpenoid catabolic process (IEA) | GO:0016115
- tetrasaccharide transport (IEA) | GO:2001098
- vacuolar acidification (IEA) | GO:0007035
- xanthophyll biosynthetic process (IEA) | GO:0016123
- xanthophyll catabolic process (IEA) | GO:0016124
 Domains
Domains
- ( PF00069 ) Protein kinase domain
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
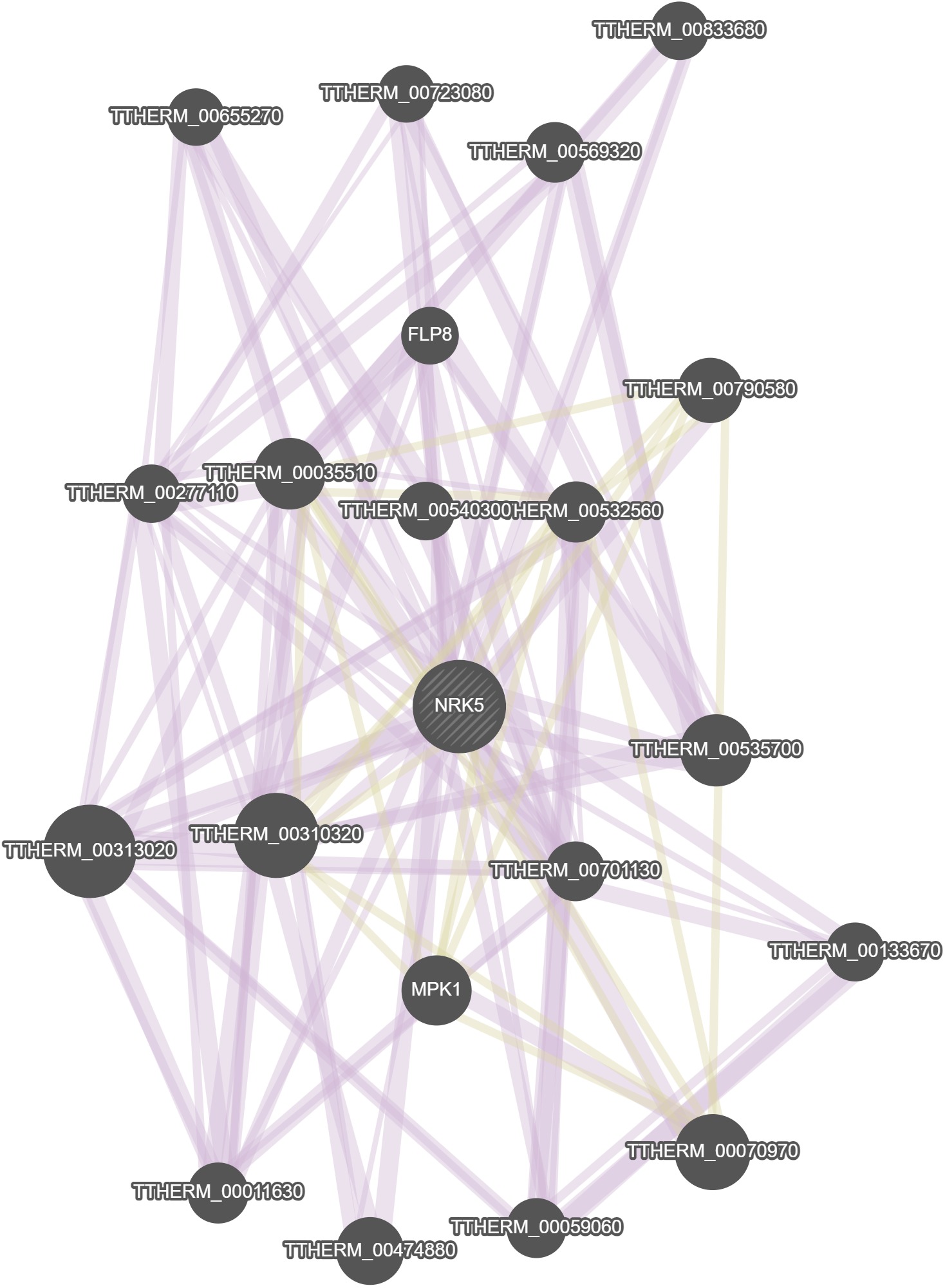
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:16611747: Wloga D, Camba A, Rogowski K, Manning G, Jerka-Dziadosz M, Gaertig J (2006) Members of the NIMA-related kinase family promote disassembly of cilia by multiple mechanisms. Molecular biology of the cell 17(6):2799-810
 Sequences
Sequences
>TTHERM_00301840(coding)
ATGGATTTACCAAACATCAATGCAAACAGTGTTTAAGATATTGGATCTCCTTCAAAAAAG
AGAGAAATATCTATAGATGAAACCGGATAGATTGGTTTAAAGAATAAATAAATAGTTTAC
AATTCGAGTAACAATAGCATGAATAAGCGTAAAAGTCCTGATTAAGTTAAAACACCTCCT
CCCCATTAAAATCATAGATCAATTAGCCTGCAACCTAATAATTAATCTGTTTTATAATCC
TCTCCATAGTAGTAATAACAAAATTAAATGGAAAAGAAAAGTAATCCTCTAAGGAGCTCT
TCGATTTTAGATGATTAATTTTTATAGCATGCATCAAATGCCTCAGCTGAAGCTGTTTAA
TAAGTGGTATCAAATTACAGAAATAATCTTATAAATTAATAATCGCAATCTCTGTAGCAG
CCACTTGTAAAGTAACATCCAAAAAATAATTCTAATAACACAAGTAGTTTACAGTAAAGT
CCTTAGCATCACTAAGGTAATACTTATTTCCATGACGAAGAACTCTAAGATTTTGAAATT
CTAAGCAAATTAGGCGAAGGTTCTTTTAGTACTGTGTATAGAGTTAGACGTGGAAAGGAT
GGAAAAGAATATGCTTTGAAAAGAATAAAGATGATGAAATTAAATGAAAAAGAAAGGGAA
AACGCAGTAAATGAAGTGAGATTTTTGGCTTCAATTAACTGCAGGAATATAATCAGCTAC
AAATAAGCTATTTATGATGAAGGAGTCAATCAATTATGTGTTATAATGGAATATGCGGAG
GGAGGCGATTTAGCTAGAATTATTCGTCATGCTTCAAAAGCAGGTAAATATATTGAGGAA
GATATGATTTGGTCATATGCTATTTAAATGACCATAGGCATTAAAGCGTTACATGATTTA
AATATATTGCATAGAGATTTAAAGGCAGCTAATGTTTTCTTAGATAAGTATTAAACAAGG
GCTATGTTGGGAGACTTGAATGTATCAAAAAAACTACAAGTGAACGGTTTACTTTATACT
CAAACTGGTACTCCTTACTATGCCTCTCCAGAAGTTTGGAAAGATAAACCTTACAATAAT
AAAAGTGATATTTGGAGTTTGGGCTGTGTTATATATGAAATGTGTGCTCTGAAACCACCA
TTTAAGGGAAAAGATATGGAAGACTTATTTAAAAAGGTATAAAGAGGTGTCTACGATCCT
ATTCCTTCTCACTTTAGCAAAGAGCTAAACCTCTTCATTGCTTAATTATTAAGGGTAAAC
CCTGAATAGCGTCCTAATTGTGATGAAATTCTCAAATTTTCTGTGATTAAAAAAAGAATG
GCTGATATGCAGCATTTCTTTTCAGATTCTCTTACTCCTTTCTAACAAAAAATGTAATCT
GAATTAATAAGCACAATTCGTATTGGGAAGTCTTTACGTGGGTTAAAAGATAAATTGCCT
CAGGCAAGATACGAAACAACGGAATCTACTTAAGCTACAACAAATTAAATAACTTCTTAG
TCAGAAATTTTACAACCTAATTAATACCACCGTCAAAGAAAAAATAAATACATTTCTCAT
ATATCGTCAAGTAATATAAGTGATATGATGAATTAACAAATAAGCATACCATCTTAACTA
TTAAATCCGTCTGTAGAAATATAAAGTAAAGGCTTGAATGATAATCACAAAGGAAGTTCA
AGTTTGAACTAGAATTCCATTTCTTTAAGATAATCTTAATAACAATAAGTTATCCCTTCT
ATATAATCATCACCTCCATCATAAAAATCTGAACATATTAAATAAATTATTGCATAAAAA
TAATAAAATAAATCTTCATTATATTAAAAATAATATTAATACCCACGGTTGCCACTTTAG
CATTAATAATTTGATAGTGCCTCTCCAATATAAGCTAAAAGCAGATGTAGTTCTGTAGAA
AGTAAGCAATCAATACATAAAAATTAGCTAAGTCTTCATAAGTAATACTACCAAAATAAT
GATATAAATAAATCTATTAATTTGAATAATTCATAAAACTCTATTGTCACAAATAAAAAA
CCGATACGAAACAAAGAAAGTAATACACCATAAATGTACTAATTATAAAAAGCAGAAGAA
CCAATATCAGTCAATTAAAGTGTCCAGTAGCTTTCAAAAGAAATAAAATAATTAAATTCT
AATTTATATAATGATGAGATGAATAGTCGCTAGAAAAAAATGTATCAACTTTAAGCTTTA
TAAAAAATCTTAGATTAAAAATGTTAACTACCTCAAATACTGAAGACATCCATATACTGA
>TTHERM_00301840(gene)
GGATAGTATAATTAATAAGTAAAATAAATACATAATTTTTTTAGCGATTTGTGAATTCTT
TCAAATGGATTTACCAAACATCAATGCAAACAGTGTTTAAGATATTGGATCTCCTTCAAA
AAAGAGAGAAATATCTATAGATGAAACCGGATAGATTGGTTTAAAGAATAAATAAATAGT
TTACAATTCGAGTAACAATAGCATGAATAAGCGTAAAAGTCCTGATTAAGTTAAAACACC
TCCTCCCCATTAAAATCATAGATCAATTAGCCTGCAACCTAATAATTAATCTGTTTTATA
ATCCTCTCCATAGTAGTAATAACAAAATTAAATGGAAAAGAAAAGTAATCCTCTAAGGAG
CTCTTCGATTTTAGATGATTAATTTTTATAGCATGCATCAAATGCCTCAGCTGAAGCTGT
TTAATAAGTGGTATCAAATTACAGAAATAATCTTATAAATTAATAATCGCAATCTCTGTA
GCAGCCACTTGTAAAGTAACATCCAAAAAATAATTCTAATAACACAAGTAGTTTACAGTA
AAGTCCTTAGCATCACTAAGGTAATACTTATTTCCATGACGAAGAACTCTAAGATTTTGA
AATTCTAAGCAAATTAGGCGAAGGTTCTTTTAGTACTGTGTATAGAGTTAGACGTGGAAA
GGATGGAAAAGAATATGCTTTGAAAAGAATAAAGATGATGAAATTAAATGAAAAAGAAAG
GGAAAACGCAGTAAATGAAGTGAGATTTTTGGCTTCAATTAACTGCAGGAATATAATCAG
CTACAAATAAGCTATTTATGATGAAGGAGTCAATCAATTATGTGTTATAATGGAATATGC
GGAGGGAGGCGATTTAGCTAGAATTATTCGTCATGCTTCAAAAGCAGGTAAATATATTGA
GGAAGATATGATTTGGTCATATGCTATTTAAATGACCATAGGCATTAAAGCGTTACATGA
TTTAAATATATTGCATAGAGATTTAAAGGCAGCTAATGTTTTCTTAGATAAGTATTAAAC
AAGGGCTATGTTGGGAGACTTGAATGTATCAAAAAAACTACAAGTGAACGGTTTACTTTA
TACTCAAACTGGTACTCCTTACTATGCCTCTCCAGAAGTTTGGAAAGATAAACCTTACAA
TAATAAAAGTGATATTTGGAGTTTGGGCTGTGTTATATATGAAATGTGTGCTCTGAAACC
ACCATTTAAGGGAAAAGATATGGAAGACTTATTTAAAAAGGTATAAAGAGGTGTCTACGA
TCCTATTCCTTCTCACTTTAGCAAAGAGCTAAACCTCTTCATTGCTTAATTATTAAGGGT
AAACCCTGAATAGCGTCCTAATTGTGATGAAATTCTCAAATTTTCTGTGATTAAAAAAAG
AATGGCTGATATGCAGCATTTCTTTTCAGATTCTCTTACTCCTTTCTAACAAAAAATGTA
ATCTGAATTAATAAGCACAATTCGTATTGGGAAGTCTTTACGTGGGTTAAAAGATAAATT
GCCTCAGGCAAGATACGAAACAACGGAATCTACTTAAGCTACAACAAATTAAATAACTTC
TTAGTCAGAAATTTTACAACCTAATTAATACCACCGTCAAAGAAAAAATAAATACATTTC
TCATATATCGTCAAGTAATATAAGTGATATGATGAATTAACAAATAAGCATACCATCTTA
ACTATTAAATCCGTCTGTAGAAATATAAAGTAAAGGCTTGAATGATAATCACAAAGGAAG
TTCAAGTTTGAACTAGAATTCCATTTCTTTAAGATAATCTTAATAACAATAAGTTATCCC
TTCTATATAATCATCACCTCCATCATAAAAATCTGAACATATTAAATAAATTATTGCATA
AAAATAATAAAATAAATCTTCATTATATTAAAAATAATATTAATACCCACGGTTGCCACT
TTAGCATTAATAATTTGATAGTGCCTCTCCAATATAAGCTAAAAGCAGATGTAGTTCTGT
AGAAAGTAAGCAATCAATACATAAAAATTAGCTAAGTCTTCATAAGTAATACTACCAAAA
TAATGATATAAATAAATCTATTAATTTGAATAATTCATAAAACTCTATTGTCACAAATAA
AAAACCGATACGAAACAAAGAAAGTAATACACCATAAATGTACTAATTATAAAAAGCAGA
AGAACCAATATCAGTCAATTAAAGTGTCCAGTAGCTTTCAAAAGAAATAAAATAATTAAA
TTCTAATTTATATAATGATGAGATGAATAGTCGCTAGAAAAAAATGTATCAACTTTAAGC
TTTATAAAAAATCTTAGATTAAAAATGTTAACTACCTCAAATACTGAAGACATCCATATA
CTGAATGATTTACAAATAAAAAACATATGTTTTTAATAAAAGATATTAATTACTTTTATT
GATTTTTACTTTAATTATTATAAAAATTAACCTTAAAAACAATTTTGTTCTAACTTAAAT
TAACTTTTAAATTCAAAATATTTTAATTAAAATCATTAAAATAATGATAAATAATTAATT
AAAGGGTATAAATTTTATAAATAGATTTATAAACTGTAAATGCTTTATCAATATTTTGTT
TATATTATATTTAATAGCTTATTTATTATATTTATTC>TTHERM_00301840(protein)
MDLPNINANSVQDIGSPSKKREISIDETGQIGLKNKQIVYNSSNNSMNKRKSPDQVKTPP
PHQNHRSISLQPNNQSVLQSSPQQQQQNQMEKKSNPLRSSSILDDQFLQHASNASAEAVQ
QVVSNYRNNLINQQSQSLQQPLVKQHPKNNSNNTSSLQQSPQHHQGNTYFHDEELQDFEI
LSKLGEGSFSTVYRVRRGKDGKEYALKRIKMMKLNEKERENAVNEVRFLASINCRNIISY
KQAIYDEGVNQLCVIMEYAEGGDLARIIRHASKAGKYIEEDMIWSYAIQMTIGIKALHDL
NILHRDLKAANVFLDKYQTRAMLGDLNVSKKLQVNGLLYTQTGTPYYASPEVWKDKPYNN
KSDIWSLGCVIYEMCALKPPFKGKDMEDLFKKVQRGVYDPIPSHFSKELNLFIAQLLRVN
PEQRPNCDEILKFSVIKKRMADMQHFFSDSLTPFQQKMQSELISTIRIGKSLRGLKDKLP
QARYETTESTQATTNQITSQSEILQPNQYHRQRKNKYISHISSSNISDMMNQQISIPSQL
LNPSVEIQSKGLNDNHKGSSSLNQNSISLRQSQQQQVIPSIQSSPPSQKSEHIKQIIAQK
QQNKSSLYQKQYQYPRLPLQHQQFDSASPIQAKSRCSSVESKQSIHKNQLSLHKQYYQNN
DINKSINLNNSQNSIVTNKKPIRNKESNTPQMYQLQKAEEPISVNQSVQQLSKEIKQLNS
NLYNDEMNSRQKKMYQLQALQKILDQKCQLPQILKTSIY