Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00313380Standard Name
CRP1 (Calcium Regulator Protein 1)Aliases
PreTt03525 | 30.m00334 | 3810.m01946Description
CRP1 sodium calcium exchanger protein; Sodium/calcium exchanger protein; Calcium:cation antiporterGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- alpha6-beta1 integrin-CD151 complex (IEA) | GO:0071059
- interleukin-20 receptor complex (IEA) | GO:0030876
- mitochondrial ATP-gated potassium channel complex (IEA) | GO:0062157
- nucleolar chromatin (IEA) | GO:0030874
- obsolete ectoplasm (IEA) | GO:0043265
- RNA polymerase complex (IEA) | GO:0030880
- subrhabdomeral cisterna (IEA) | GO:0016029
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 3-hydroxybutyrate dehydrogenase activity (IEA) | GO:0003858
- amino acid transmembrane transporter activity (IEA) | GO:0015171
- aminotriazole transporter activity (IEA) | GO:0015241
- arabinogalactan binding (IEA) | GO:2001085
- arsenate reductase (glutaredoxin) activity (IEA) | GO:0008794
- carboxymethylenebutenolidase activity (IEA) | GO:0008806
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- fucosyltransferase activity (IEA) | GO:0008417
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- gamma-aminobutyric acid transmembrane transporter activity (IEA) | GO:0015185
- hercynylcysteine sulfoxide lyase activity (ergothioneine-forming) (IEA) | GO:1990411
- high-affinity ferric iron transmembrane transporter activity (IEA) | GO:0015092
- inositol hexakisphosphate 2-phosphatase activity (IEA) | GO:0052826
- L-aspartate transmembrane transporter activity (IEA) | GO:0015183
- L-cystine transmembrane transporter activity (IEA) | GO:0015184
- L-glutamine transmembrane transporter activity (IEA) | GO:0015186
- lipopolysaccharide heptosyltransferase activity (IEA) | GO:0008920
- magnesium ion transmembrane transporter activity (IEA) | GO:0015095
- melibiose:monoatomic cation symporter activity (IEA) | GO:0015487
- microtubule severing ATPase activity (IEA) | GO:0008568
- multidrug efflux pump activity (IEA) | GO:0015559
- myosin III binding (IEA) | GO:0031473
- O-phospho-L-serine:2-oxoglutarate aminotransferase activity (IEA) | GO:0004648
- obsolete anticoagulant activity (IEA) | GO:0008435
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete N2-acetyl-L-aminoadipate semialdehyde dehydrogenase activity (IEA) | GO:0043745
- obsolete phosphatidylinositol 3-kinase, class I, catalyst activity (IEA) | GO:0015072
- obsolete Rieske iron-sulfur protein (IEA) | GO:0000047
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete zinc, iron permease activity (IEA) | GO:0015342
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoribosyl-AMP cyclohydrolase activity (IEA) | GO:0004635
- polyamine transmembrane transporter activity (IEA) | GO:0015203
- protein-N-terminal asparagine amidohydrolase activity (IEA) | GO:0008418
- purinergic nucleotide receptor activity (IEA) | GO:0001614
- pyrimidine nucleotide-sugar transmembrane transporter activity (IEA) | GO:0015165
- RNA lariat debranching enzyme activity (IEA) | GO:0008419
- RNA polymerase II CTD heptapeptide repeat phosphatase activity (IEA) | GO:0008420
- Ser(Gly)-tRNA(Ala) hydrolase activity (IEA) | GO:0043908
- sulfite oxidase activity (IEA) | GO:0008482
Biological Process
- aspartate biosynthetic process (IEA) | GO:0006532
- cadmium ion transport (IEA) | GO:0015691
- cell adhesion involved in multi-species biofilm formation (IEA) | GO:0043710
- cellotriose transport (IEA) | GO:2001096
- cellular response to desiccation (IEA) | GO:0071465
- cellular response to magnesium ion (IEA) | GO:0071286
- coenzyme A biosynthetic process (IEA) | GO:0015937
- DNA recombinase mediator complex assembly (IEA) | GO:1903871
- extraembryonic membrane development (IEA) | GO:1903867
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- gastrulation involving germ band extension (IEA) | GO:0010004
- glycolytic process from glycerol (IEA) | GO:0061613
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- homogentisate metabolic process (IEA) | GO:1901999
- laminaritriose transport (IEA) | GO:2001097
- loss of chromatin silencing (IEA) | GO:0006345
- lung connective tissue development (IEA) | GO:0060427
- lung saccule development (IEA) | GO:0060430
- maltoheptaose metabolic process (IEA) | GO:2001122
- methanol metabolic process (IEA) | GO:0015945
- mitotic nuclear division (IEA) | GO:0007067
- modulation of formation of symbiont germ tube hook structure for appressorium development (IEA) | GO:0075030
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phospholipid translocation (IEA) | GO:0061093
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- obsolete aging (IEA) | GO:0007568
- obsolete induction by virus of host cell-cell fusion (IEA) | GO:0006948
- obsolete modulation by symbiont of defense-related host calcium-dependent protein kinase pathway (IEA) | GO:0052168
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete reproduction (IEA) | GO:0000003
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- phenanthrene catabolic process via trans-9(R),10(R)-dihydrodiolphenanthrene (IEA) | GO:0018956
- phenanthrene catabolic process via trans-9(S),10(S)-dihydrodiolphenanthrene (IEA) | GO:0018957
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- primitive palate development (IEA) | GO:0062010
- process resulting in tolerance to ketone (IEA) | GO:1990369
- programmed cell death (IEA) | GO:0012501
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- prostate induction (IEA) | GO:0060514
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of antisense RNA transcription (IEA) | GO:0060194
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of frizzled signaling pathway (IEA) | GO:0008590
- regulation of interferon-beta secretion (IEA) | GO:0035547
- regulation of phosphatidylcholine biosynthetic process (IEA) | GO:2001245
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of satellite cell activation involved in skeletal muscle regeneration (IEA) | GO:0014717
- regulation of serine-type peptidase activity (IEA) | GO:1902571
- rRNA pseudouridine synthesis (IEA) | GO:0031118
- sophorose transport (IEA) | GO:2001087
- sperm aster formation (IEA) | GO:0035044
- sperm DNA decondensation (IEA) | GO:0035041
- symbiont-mediated suppression of host innate immune response (IEA) | GO:0052170
- syncytium formation (IEA) | GO:0006949
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- tetrasaccharide transport (IEA) | GO:2001098
- thyroid gland development (IEA) | GO:0030878
- trisaccharide transport (IEA) | GO:2001088
- xylotriose transport (IEA) | GO:2001094
 Domains
Domains
- ( PF01699 ) Sodium/calcium exchanger protein
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
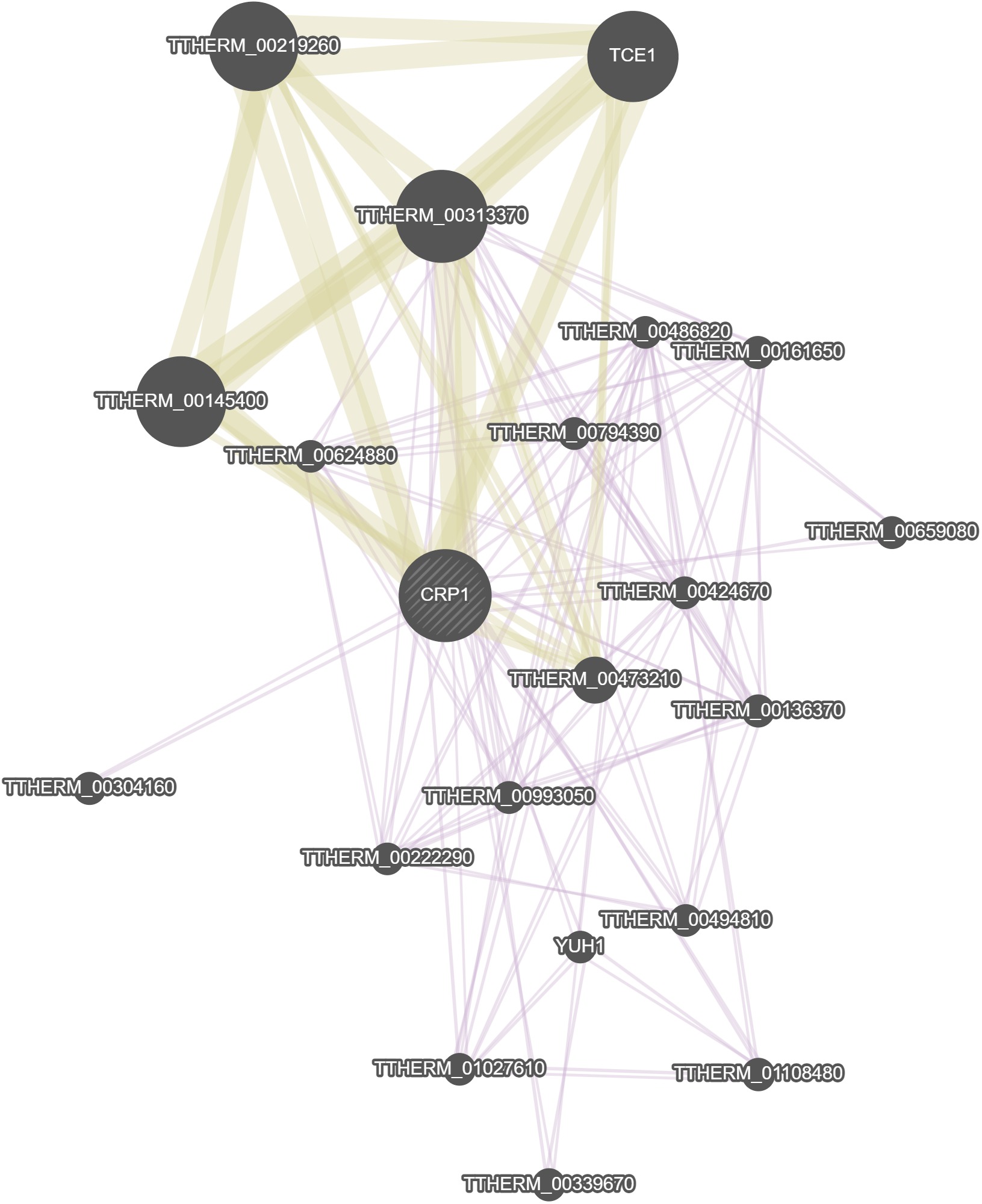
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No. Gene Name(s) Paragraph Text 2258 CRP1 Tetrahymena thermophilia CRP1 (calcium regulator protein 1) appears to be a member of the sodium/calcium exchanger protein family. It contains one putative sodium/calcium exchanger domain, which has an E-value of 3.2*10^-13, indicating that it is reasonably related to this domain sequence. It is possible that this protein regulates Ca2+ cation concentration within the cell, where the movement of Ca2+ in or out of the cytoplasm is contingent upon the concentration of Na+ in the cell. M. Huen and M. Greeley
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00313380(coding)
ATGGCAGACTAATCTTAAATGTGTACTTTTAAAAAAATACAGGCATTGCAATAGCAAGAT
ATCTGTGCTTACGCAGATCAGCATTGCAATGTTGAGTAAATGATAAGATTTTCTTACTTT
TACTATTGCTAAGTCAATTAAAATATTCTTGTTTTAGACCTTCTTACAATATTGGTTCCA
TTTATTTCCTTCCGTTTACTAAGCTAAACATCTGAATACTACCTTTCCCCTTCATTAGCT
AAGCTAGCTAAGTGCTTTAGACTCTCTCAAACCTTGGCAGGCGTAACATTACTTGCCTTA
GGAAATGGTGGTCCTGATGTGATCACGGCAATTATTGCAGGAAACAGTGGAGATGATTCT
ATTTCTATAGCAGTTGGATCTATATTTGGCGCAGGGCTATTTGTGACAACTTACACTCTT
GCTAATTGTATTCAGAACGCCGGTTAAATCAAGATTCAATCTAGAACTTTTTTTAGAGAT
CAACTATTCTATTTAATTGGATGTTTGGTTATTCTCATCTACACAATAATAGGTGAAGTA
ACTTTAAGCATGTCAATTGGTTTTATAAGTATCTACATAATATTCTTGCTAGTTGTAGTC
TATTAAGATAAGACTAATCCACAAAAAAAAGAATCAGAGGATAGCACTAAGCAACAATAA
ACTATCAGAAAAATGGAAACAATGCAAAGTAATTTTTTCTCCTCTAATTCCATCAACACT
TTAACAATTTATACAGCTTAAAAGATAGATGACATTCAAATCGAAATGTAAATTGAATAA
ATGAGAAATGACCCCATGGCTATGATGCATAGACAAACAACTCTTGCTGAATTTAGATAA
TCAGATCTCCCTCTTTGCGATCTAAAAGAGTCGAACGATTAAGACATGAAGGATCAAGAA
AACAAAGTTCCAAGCATCCCCATGATAATTATCCCCGAAGATTAAGAAGAAGATGAAGAA
AATGGGATTAGAAGCAAAAAATCTTCTTTAGAAGAATAATAAAAATAATAGTAGAACACT
GAAGCTTTTGATACTCTAAAGAGATGCGATACTTAAAATTAAGCTCAAACTTATGAGTAA
GCTGACGATGAATCAAGTAGTAAATCATCTAATGACTCTAATAACAAAGATAGCATGACT
GAAAAGTACGAAAACTTCAAGTAAAAGCTTCATGAAATGTCTGTCCCTGAAAAAATCTTA
GCTTTAGCTGAAATTCCTATTGACCTTATTAGATACTTCATCATCCCTCCAGCTGAAGAT
TCTTAATGGAATAAAATAAGAGCGATTGTTTGTTGCTACACTTCTCCAATCGTGTTCCTC
CTCTGCTTTGGAGGTATAACAATGCAAATTAATGGAGTTGAAGGATTTTATGTTGCTTAC
CTTCTTCTTGCTATTTCTTTTGTATTATCAATTATTGTTTTCTTCTTTACCAAGAAAGAC
TAAGCTCCTTGTTTCCAATGGCTTTTTTCCTTAGGCTCAATATTTGTCGCTATTGCAATT
CTTAGTGTTATAGCCTAAGTCTTAGTAGATTTTATTAACTTTTTCTAGCTTCTCACATCA
ATGAATAAAACTTTCTTGGGTATGACTCTTTTAGCCTTAGGAAACTCTGCTACAGATTTC
TTTACTAATTCATAACTTGCCAAAGTTGGATATGGTGTTATGGCTTTAACTGGATGCTTT
GCTGGACAAGCATTTGATTTATACCTAGGATTCGGTTTTGCTTTAGTTTTTAGCTCTTAT
AAACAAGGATTTGGTTTATCTATTTTCACAGGCTCTAACATCAATGACTGGTTTAGTAAC
AGTATTGCTATTACAATCTTAGTTTCTGTGATTATTTCAACCATAATTATACTCATATGT
TCCATAAAACAAAATTACAATTTTGAAAAAGGAAAATTCACCAGCTTTATCAAAAATTAC
TACTTACTCATAATTACTATCTGCATAGGTCTCTGTGCAGCTGACTAATTCTTTGAATGA
>TTHERM_00313380(gene)
AAATTAATTAACAAAATTGAAAAAAAAAAAAACAAATAGAGAGAATTTAAAAAAAAGAAA
AACAAAATAAATAAAAAATTAAAATGGCAGACTAATCTTAAATGTGTACTTTTAAAAAAA
TACAGGCATTGCAATAGCAAGATATCTGTGCTTACGCAGATCAGCATTGCAATGTTGAGT
AAATGATAAGATTTTCTTACTTTTACTATTGCTAAGTCAATTAAAATATTCTTGTTTTAG
ACCTTCTTACAATATTGGTTCCATTTATTTCCTTCCGTTTACTAAGCTAAACATCTGAAT
ACTACCTTTCCCCTTCATTAGCTAAGCTAGCTAAGTGCTTTAGACTCTCTCAAACCTTGG
CAGGCGTAACATTACTTGCCTTAGGAAATGGTGGTCCTGATGTGATCACGGCAATTATTG
CAGGAAACAGTGGAGATGATTCTATTTCTATAGCAGTTGGATCTATATTTGGCGCAGGGC
TATTTGTGACAACTTACACTCTTGCTAATTGTATTCAGAACGCCGGTTAAATCAAGATTC
AATCTAGAACTTTTTTTAGAGATCAACTATTCTATTTAATTGGATGTTTGGTTATTCTCA
TCTACACAATAATAGGTGAAGTAACTTTAAGCATGTCAATTGGTTTTATAAGTATCTACA
TAATATTCTTGCTAGTTGTAGTCTATTAAGATAAGACTAATCCACAAAAAAAAGAATCAG
AGGATAGCACTAAGCAACAATAAACTATCAGAAAAATGGAAACAATGCAAAGTAATTTTT
TCTCCTCTAATTCCATCAACACTTTAACAATTTATACAGCTTAAAAGATAGATGACATTC
AAATCGAAATGTAAATTGAATAAATGAGAAATGACCCCATGGCTATGATGCATAGACAAA
CAACTCTTGCTGAATTTAGATAATCAGATCTCCCTCTTTGCGATCTAAAAGAGTCGAACG
ATTAAGACATGAAGGATCAAGAAAACAAAGTTCCAAGCATCCCCATGATAATTATCCCCG
AAGATTAAGAAGAAGATGAAGAAAATGGGATTAGAAGCAAAAAATCTTCTTTAGAAGAAT
AATAAAAATAATAGTAGAACACTGAAGCTTTTGATACTCTAAAGAGATGCGATACTTAAA
ATTAAGCTCAAACTTATGAGTAAGCTGACGATGAATCAAGTAGTAAATCATCTAATGACT
CTAATAACAAAGATAGCATGACTGAAAAGTACGAAAACTTCAAGTAAAAGCTTCATGAAA
TGTCTGTCCCTGAAAAAATCTTAGCTTTAGCTGAAATTCCTATTGACCTTATTAGATACT
TCATCATCCCTCCAGCTGAAGATTCTTAATGGAATAAAATAAGAGCGATTGTTTGTTGCT
ACACTTCTCCAATCGTGTTCCTCCTCTGCTTTGGAGGTATAACAATGCAAATTAATGGAG
TTGAAGGATTTTATGTTGCTTACCTTCTTCTTGCTATTTCTTTTGTATTATCAATTATTG
TTTTCTTCTTTACCAAGAAAGACTAAGCTCCTTGTTTCCAATGGCTTTTTTCCTTAGGCT
CAATATTTGTCGCTATTGCAATTCTTAGTGTTATAGCCTAAGTCTTAGTAGATTTTATTA
ACTTTTTCTAGCTTCTCACATCAATGAATAAAACTTTCTTGGGTATGACTCTTTTAGCCT
TAGGAAACTCTGCTACAGATTTCTTTACTAATTCATAACTTGCCAAAGTTGGATATGGTG
TTATGGCTTTAACTGGATGCTTTGCTGGACAAGCATTTGATTTATACCTAGGATTCGGTT
TTGCTTTAGTTTTTAGCTCTTATAAACAAGGATTTGGTTTATCTATTTTCACAGGCTCTA
ACATCAATGACTGGTTTAGTAACAGTATTGCTATTACAATCTTAGTTTCTGTGATTATTT
CAACCATAATTATACTCATATGTTCCATAAAACAAAATTACAATTTTGAAAAAGGAAAAT
TCACCAGCTTTATCAAAAATTACTACTTACTCATAATTACTATCTGCATAGGTCTCTGTG
CAGCTGACTAATTCTTTGAATGAAATACTTATTACTATTTCACTGTCACTATTTAATAAC
ACTCTCACATACTTTCTAACTGTCAATTTACTTAAAACTTTTCTTAAATAAATATCTTTC
GATATTTAAACAAACCTATATTAATATAAAAATATAAAATAATAATTAATATCTACTATA
TAAATTTATTTATATAGTTTTAGTCTTGTATTCTTTCTTATAAAAAAATTATATCTTAGT
ATTTTCTTCTGAAAATTCATGCATTCTTTCACTCCTACTAAATCCACAAATAAATGCGTA
AATTAAATTA>TTHERM_00313380(protein)
MADQSQMCTFKKIQALQQQDICAYADQHCNVEQMIRFSYFYYCQVNQNILVLDLLTILVP
FISFRLLSQTSEYYLSPSLAKLAKCFRLSQTLAGVTLLALGNGGPDVITAIIAGNSGDDS
ISIAVGSIFGAGLFVTTYTLANCIQNAGQIKIQSRTFFRDQLFYLIGCLVILIYTIIGEV
TLSMSIGFISIYIIFLLVVVYQDKTNPQKKESEDSTKQQQTIRKMETMQSNFFSSNSINT
LTIYTAQKIDDIQIEMQIEQMRNDPMAMMHRQTTLAEFRQSDLPLCDLKESNDQDMKDQE
NKVPSIPMIIIPEDQEEDEENGIRSKKSSLEEQQKQQQNTEAFDTLKRCDTQNQAQTYEQ
ADDESSSKSSNDSNNKDSMTEKYENFKQKLHEMSVPEKILALAEIPIDLIRYFIIPPAED
SQWNKIRAIVCCYTSPIVFLLCFGGITMQINGVEGFYVAYLLLAISFVLSIIVFFFTKKD
QAPCFQWLFSLGSIFVAIAILSVIAQVLVDFINFFQLLTSMNKTFLGMTLLALGNSATDF
FTNSQLAKVGYGVMALTGCFAGQAFDLYLGFGFALVFSSYKQGFGLSIFTGSNINDWFSN
SIAITILVSVIISTIIILICSIKQNYNFEKGKFTSFIKNYYLLIITICIGLCAADQFFE