Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00333210Standard Name
RPL4 (Ribosomal Protein of the large subunit)Aliases
PreTt05965 | 34.m00202 | 3689.m00001Description
RPL4 60S ribosomal protein L4; ribosomal protein L4/L1 family protein; Large ribosomal subunit protein uL4, eukaryotic and archaeal typeGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 11beta-hydroxyprogesterone binding (IEA) | GO:1903879
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- CCR10 chemokine receptor binding (IEA) | GO:0031735
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- prenyltransferase activity (IEA) | GO:0004659
- protein histidine kinase activity (IEA) | GO:0004673
Biological Process
- asymmetric protein localization (IEA) | GO:0008105
- basement membrane organization (IEA) | GO:0071711
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- coenzyme A metabolic process (IEA) | GO:0015936
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- microtubule depolymerization (IEA) | GO:0007019
- mitochondrial membrane organization (IEA) | GO:0007006
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of glycoprotein metabolic process (IEA) | GO:1903019
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- negative regulation of stomatal opening (IEA) | GO:1902457
- negative regulation of telomere single strand break repair (IEA) | GO:1903824
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete bent DNA binding (IEA) | GO:0006326
- obsolete larval behavior (sensu Insecta) (IEA) | GO:0007627
- obsolete MAPK cascade involved in ascospore formation (IEA) | GO:1903695
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- pigment accumulation in response to UV light (IEA) | GO:0043478
- pigment accumulation in tissues in response to UV light (IEA) | GO:0043479
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of B cell mediated immunity (IEA) | GO:0002714
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of peroxisome proliferator activated receptor signaling pathway (IEA) | GO:0035360
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903691
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of glycoprotein metabolic process (IEA) | GO:1903018
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
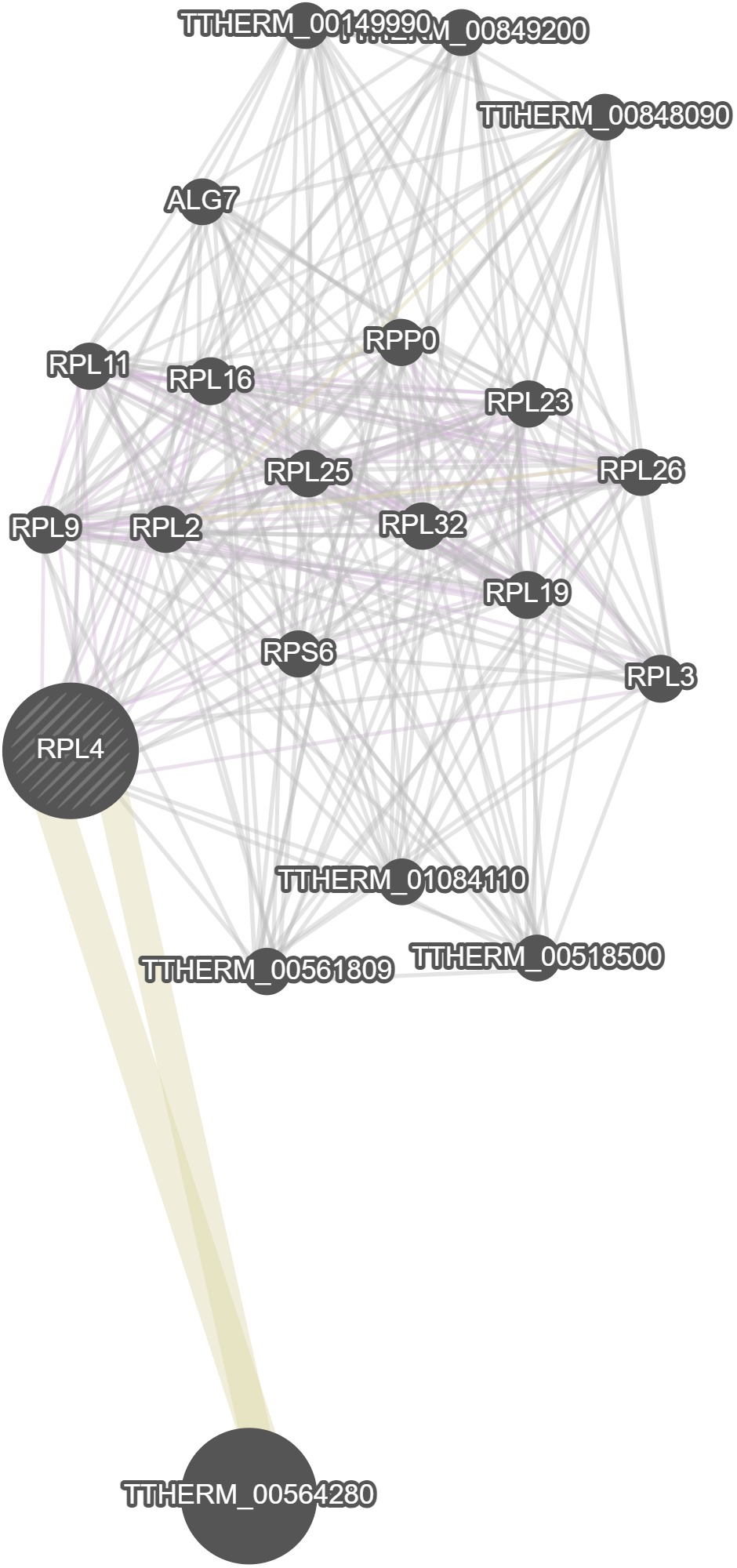
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:22052974: Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N (2011) Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science (New York, N.Y.) 334(6058):941-8
 Sequences
Sequences
>TTHERM_00333210(coding)
ATGACTTCTAGACCTCAAATCCACGTCCACGACGCTAAGGAAGCTAACAAGCAAACCGCT
ACCAAGCTCACCCTCCCCGCTGTCTTCACTGCTCCCATCAGAACTGATATTGTCCACAAG
GTTTTCACCGACCTTAACAAGAACAGAAAGCAAGCTTCCGGAGTCAAGATTTCCACCAGA
GGTACTGCTGGTATGGGTCACTCCGCTGAATCTTGGGGTACTGGTAGAGCTGTTGCTCGT
ATTCCCAGAGTTGGTGGTTCTGGTACTCACAGATCCGGTCAAGCCGCCTTCGGTAACCAA
TGTAGAAAGGGTAGAATGTTCGCTCCCTTGAAGACCTACAGAAGAGTCCACAGAAGAGTC
AATGTCAACCAAAAGAGACACGCCGTCGCTGCTGCCCTCGCTGCCTCTGCTTTGGTTCCT
TTAGTTTTCGCTAGAGGTCACAGAATCTCCAACGTTTAGGAACTCCCCTACGTTTTCGAT
GACTCCGTCGAATCTTATGAAAAGACCAAGCAAGCTGTTGCTTTCCTCAAGAGAGTCGGT
GCTTACGATGATGTCCTCAGAGTTGCTGAAACCAAGGCACTCAGAGCTGGTCAAGGTAAA
TTGAGAAACAGAAGATACAAACTCAGAAGAGGTCCCCTCGTCGTCTACGGTAACGAAAAG
TCCACCCTCACCAGAGCTCTTAGAAACATTCCTGGTGTTGATGTTTGCAACGTTAACAGA
CTTAACTTATTATAATTGGCTCCTGGTGGTCACGTTGGTAGATTCATCATCTGGACTGAA
TCTGCTTTCAAGAAGTTAAACGAAATCTTCGGTACTTACTCTACTACTGGTGTCCAAAAG
AGCGGTTACCAATTATAAAGACCCCTCTTGGCCAACGCTGACATCGCTAGAATCATCAAC
TCTAACGAAGTCTAATCCGTTGTTAAGGTTGCTGGTACCACCGAAACCCACGAAAGAAAG
AAAAACCCCTTGACCAACAACAACGCTCTCTTCAAGTTAAACCCCGCTGCTAAGATCGTT
AAGGAACAAGCTAAGAAGGCTGCTGAAGCTTCCAAGGCTAAGAGATAAGCTACCCTCAAG
GCCAACAGAAAGGCTGCCAAGACCCACAAGAAGGGATCTCAAGCTTGGATTGCTGCTTTC
AACAAGGCTAACGAAGAAGCTATTGCTAAGGCTAGATAAGAAGATGCTGATTTCATTGCT
CAAGGTCAAGAAATCAAGGAAGGTGACGAATGA>TTHERM_00333210(gene)
TTTAAAGAGGGGGAAACATATTGATTTTAAGGCCAAATCAAAATATCTATTAAGAGAAAT
TAATCAAAGGGAAAAACAAATAAAAGTAAAGTAAAAGATAAATTAAAAAGTATTAGAAAA
AAAAAATAAAAGAAATGACTTCTAGACCTCAAATCCACGTCCACGACGCTAAGGAAGCTA
ACAAGCAAACCGCTACCAAGCTCACCCTCCCCGCTGTCTTCACTGCTCCCATCAGAACTG
ATATTGTCCACAAGGTTTTCACCGACCTTAACAAGAACAGAAAGCAAGCTTCCGGAGTCA
AGATTTCCACCAGAGGTACTGCTGGTATGGGTCACTCCGCTGAATCTTGGGGTACTGGTA
GAGCTGTTGCTCGTATTCCCAGAGTTGGTGGTTCTGGTACTCACAGATCCGGTCAAGCCG
CCTTCGGTAACCAATGTAGAAAGGGTAGAATGTTCGCTCCCTTGAAGACCTACAGAAGAG
TCCACAGAAGAGTCAATGTCAACCAAAAGAGACACGCCGTCGCTGCTGCCCTCGCTGCCT
CTGCTTTGGTTCCTTTAGTTTTCGCTAGAGGTCACAGAATCTCCAACGTTTAGGAACTCC
CCTACGTTTTCGATGACTCCGTCGAATCTTATGAAAAGACCAAGCAAGCTGTTGCTTTCC
TCAAGAGAGTCGGTGCTTACGATGATGTCCTCAGAGTTGCTGAAACCAAGGCACTCAGAG
CTGGTCAAGGTAAATTGAGAAACAGAAGATACAAACTCAGAAGAGGTCCCCTCGTCGTCT
ACGGTAACGAAAAGTCCACCCTCACCAGAGCTCTTAGAAACATTCCTGGTGTTGATGTTT
GCAACGTTAACAGACTTAACTTATTATAATTGGCTCCTGGTGGTCACGTTGGTAGATTCA
TCATCTGGACTGAATCTGCTTTCAAGAAGTTAAACGAAATCTTCGGTACTTACTCTACTA
CTGGTGTCCAAAAGAGCGGTTACCAATTATAAAGACCCCTCTTGGCCAACGCTGACATCG
CTAGAATCATCAACTCTAACGAAGTCTAATCCGTTGTTAAGGTTGCTGGTACCACCGAAA
CCCACGAAAGAAAGAAAAACCCCTTGACCAACAACAACGCTCTCTTCAAGTTAAACCCCG
CTGCTAAGATCGTTAAGGAACAAGCTAAGAAGGCTGCTGAAGCTTCCAAGGCTAAGAGAT
AAGCTACCCTCAAGGCCAACAGAAAGGCTGCCAAGACCCACAAGAAGGGATCTCAAGCTT
GGATTGCTGCTTTCAACAAGGCTAACGAAGAAGCTATTGCTAAGGCTAGATAAGAAGATG
CTGATTTCATTGCTCAAGGTCAAGAAATCAAGGAAGGTGACGAATGAACGTGATCTATAA
TCTGAATTTTAAACCTCTTTATCTAATAAACTGCACATAAAACTATAAATATATATATAT
TCACTTAATCATACGCTATTTAATATTGTATTAAATATTTTATGGTAACCTACTTCGTAG
GATATTTAAATTTCC>TTHERM_00333210(protein)
MTSRPQIHVHDAKEANKQTATKLTLPAVFTAPIRTDIVHKVFTDLNKNRKQASGVKISTR
GTAGMGHSAESWGTGRAVARIPRVGGSGTHRSGQAAFGNQCRKGRMFAPLKTYRRVHRRV
NVNQKRHAVAAALAASALVPLVFARGHRISNVQELPYVFDDSVESYEKTKQAVAFLKRVG
AYDDVLRVAETKALRAGQGKLRNRRYKLRRGPLVVYGNEKSTLTRALRNIPGVDVCNVNR
LNLLQLAPGGHVGRFIIWTESAFKKLNEIFGTYSTTGVQKSGYQLQRPLLANADIARIIN
SNEVQSVVKVAGTTETHERKKNPLTNNNALFKLNPAAKIVKEQAKKAAEASKAKRQATLK
ANRKAAKTHKKGSQAWIAAFNKANEEAIAKARQEDADFIAQGQEIKEGDE