Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00338460Standard Name
PFK2 (PhosphoFructoKinase)Aliases
PreTt06086 | 34.m00323 | 3689.m00119Description
PFK2 6-phosphofructokinase; Beta subunit of heterooctameric Phosphofructokinase; Phosphofructokinase type AGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- aleurone grain membrane (IEA) | GO:0032578
- CURI complex (IEA) | GO:0032545
- Hpa2 acetyltransferase complex (IEA) | GO:1990331
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- upper tip-link density (IEA) | GO:1990435
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- adenylate cyclase inhibitor activity (IEA) | GO:0010855
- adenylate cyclase regulator activity (IEA) | GO:0010854
- alpha-humulene synthase activity (IEA) | GO:0080017
- aromatase activity (IEA) | GO:0008402
- aspartate-tRNA ligase activity (IEA) | GO:0004815
- cyclase activator activity (IEA) | GO:0010853
- cyclase inhibitor activity (IEA) | GO:0010852
- gamma-butyrobetaine dioxygenase activity (IEA) | GO:0008336
- nociceptin receptor binding (IEA) | GO:0031853
- obsolete histone threonine kinase activity (IEA) | GO:0035184
- obsolete promoter binding (IEA) | GO:0010843
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- oligoxyloglucan reducing-end-specific cellobiohydrolase activity (IEA) | GO:0033945
- orexigenic neuropeptide QRFP receptor binding (IEA) | GO:0031854
- oxidative RNA demethylase activity (IEA) | GO:0035515
- peptide-glutamate-alpha-N-acetyltransferase activity (IEA) | GO:1990190
- phosphatidylcholine:cardiolipin O-linoleoyltransferase activity (IEA) | GO:0032577
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- Pup ligase activity (IEA) | GO:0061664
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- sterol 14-demethylase activity (IEA) | GO:0008398
- UFM1 ligase activity (IEA) | GO:0061666
Biological Process
- acute-phase response (IEA) | GO:0006953
- alkaloid biosynthetic process (IEA) | GO:0009821
- amphisome-lysosome fusion (IEA) | GO:0061911
- asymmetric cell division (IEA) | GO:0008356
- autoinducer AI-2 transmembrane transport (IEA) | GO:1905887
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to hydroxyurea (IEA) | GO:0072711
- cellular response to osmotic stress (IEA) | GO:0071470
- determination of left/right symmetry (IEA) | GO:0007368
- ergot alkaloid biosynthetic process (IEA) | GO:0035837
- establishment of tissue polarity (IEA) | GO:0007164
- evoked neurotransmitter secretion (IEA) | GO:0061670
- external genitalia morphogenesis (IEA) | GO:0035261
- female germline ring canal stabilization (IEA) | GO:0008335
- gastrulation (IEA) | GO:0007369
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- internal genitalia morphogenesis (IEA) | GO:0035260
- larval burrowing behavior (IEA) | GO:0035181
- macrophage derived foam cell differentiation (IEA) | GO:0010742
- mannosyl-inositol phosphorylceramide metabolic process (IEA) | GO:0006675
- maternal determination of anterior/posterior axis, embryo (IEA) | GO:0008358
- microtubule cytoskeleton organization involved in mitosis (IEA) | GO:1902850
- multicellular organism development (IEA) | GO:0007275
- negative regulation of calcium ion import across plasma membrane (IEA) | GO:1905949
- negative regulation of cellular response to oxidopamine (IEA) | GO:1905847
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of D-glucose transmembrane transport (IEA) | GO:0010829
- negative regulation of hemopoiesis (IEA) | GO:1903707
- negative regulation of keratinocyte proliferation (IEA) | GO:0010839
- negative regulation of lipid storage (IEA) | GO:0010888
- negative regulation of meiotic joint molecule formation (IEA) | GO:0010947
- negative regulation of myotube differentiation (IEA) | GO:0010832
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- negative regulation of tumor necrosis factor-mediated signaling pathway (IEA) | GO:0010804
- neuropeptide catabolic process (IEA) | GO:0010813
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete acetate catabolic process to butyrate, ethanol, acetone and butanol (IEA) | GO:0010123
- obsolete activation of MAPK activity (IEA) | GO:0000187
- obsolete histone H4-K20 demethylation (IEA) | GO:0035574
- obsolete MAPK cascade involved in ascospore formation (IEA) | GO:1903695
- obsolete multi-organism cellular localization (IEA) | GO:1902581
- obsolete negative regulation of histone H3-K79 dimethylation (IEA) | GO:1905472
- obsolete negative regulation of response to extracellular stimulus (IEA) | GO:0032105
- obsolete positive regulation of mitochondrion organization (IEA) | GO:0010822
- obsolete positive regulation of mitotic G1 cell cycle arrest in response to nitrogen starvation (IEA) | GO:1903694
- obsolete regulation of apurinic/apyrimidinic endodeoxyribonuclease activity (IEA) | GO:0052721
- obsolete regulation of cholesterol esterification (IEA) | GO:0010872
- obsolete regulation of chromatin disassembly (IEA) | GO:0010848
- obsolete regulation of glucose-6-phosphatase activity (IEA) | GO:0032114
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of cell-substrate adhesion (IEA) | GO:0010811
- positive regulation of cholesterol storage (IEA) | GO:0010886
- positive regulation of D-glucose transmembrane transport (IEA) | GO:0010828
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of mitochondrial translation in response to stress (IEA) | GO:0010892
- positive regulation of myotube differentiation (IEA) | GO:0010831
- positive regulation of pancreatic amylase secretion by cholecystokinin signaling pathway (IEA) | GO:1902279
- positive regulation of Rac protein signal transduction (IEA) | GO:0035022
- positive regulation of siRNA processing (IEA) | GO:1903705
- positive regulation of steroid biosynthetic process (IEA) | GO:0010893
- preblastoderm mitotic cell cycle (IEA) | GO:0035185
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of beta-glucan metabolic process (IEA) | GO:0032950
- regulation of border follicle cell migration (IEA) | GO:1903684
- regulation of cell size (IEA) | GO:0008361
- regulation of cell-substrate adhesion (IEA) | GO:0010810
- regulation of cellular response to gamma radiation (IEA) | GO:1905843
- regulation of chitin metabolic process (IEA) | GO:0032882
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of dolichol biosynthetic process (IEA) | GO:0010794
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of keratinocyte proliferation (IEA) | GO:0010837
- regulation of meiotic joint molecule formation (IEA) | GO:0010946
- regulation of protein localization to membrane (IEA) | GO:1905475
- regulation of sprouting angiogenesis (IEA) | GO:1903670
- regulation of T cell chemotaxis (IEA) | GO:0010819
- regulation of triglyceride biosynthetic process (IEA) | GO:0010866
- response to ketamine (IEA) | GO:1901986
- single-organism intracellular transport (IEA) | GO:1902582
- sodium-independent prostaglandin transport (IEA) | GO:0071720
- sodium-independent thromboxane transport (IEA) | GO:0071721
- spermatid nucleus differentiation (IEA) | GO:0007289
- spermatogenesis (IEA) | GO:0007283
- spermatogonial cell division (IEA) | GO:0007284
- substance P catabolic process (IEA) | GO:0010814
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- ventral furrow formation (IEA) | GO:0007370
- vesicle cargo loading (IEA) | GO:0035459
 Domains
Domains
- ( PF00365 ) Phosphofructokinase
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
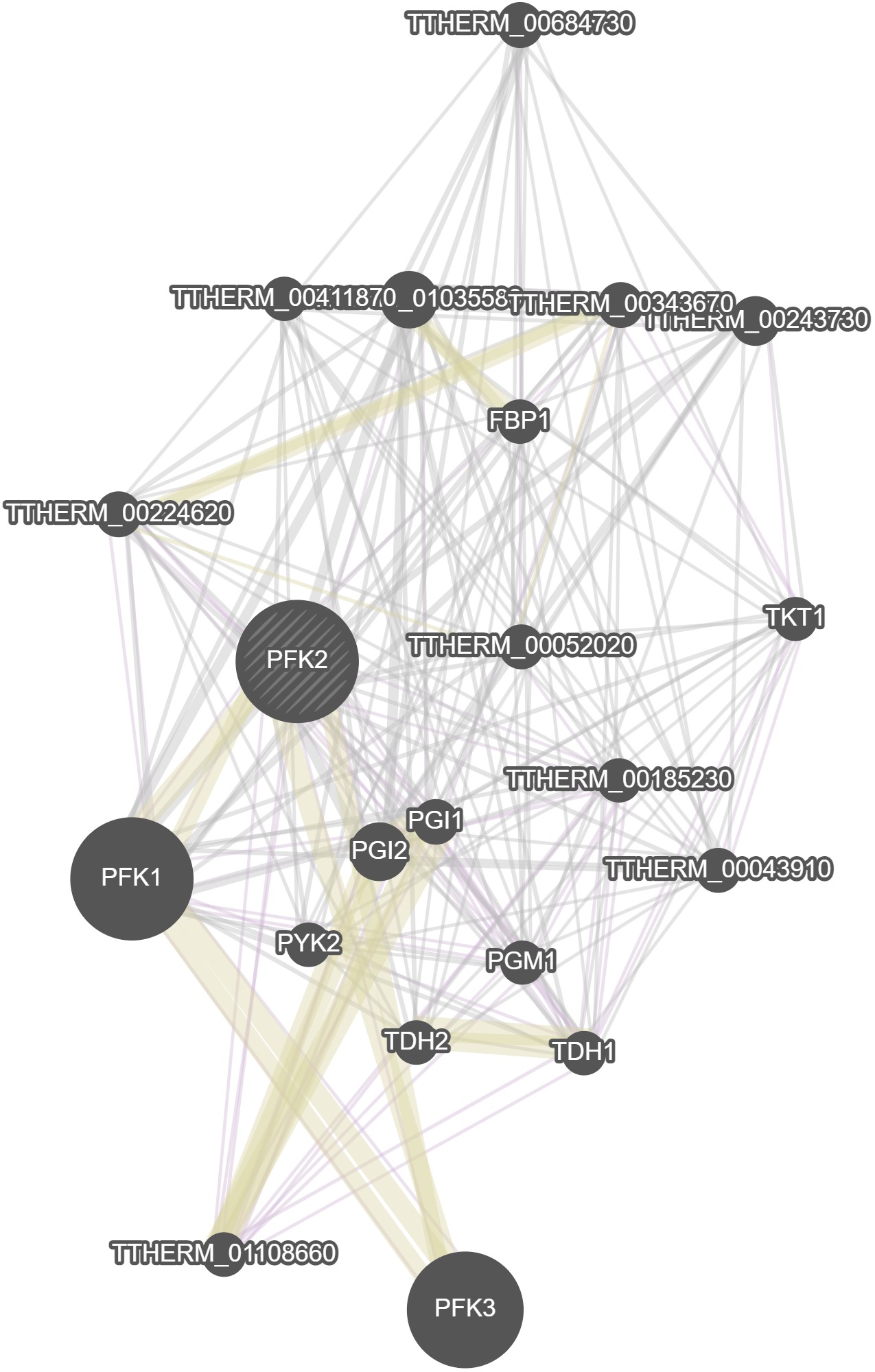
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00338460(coding)
ATGGAAAAAAACACTGTTTTGACTACAAGCATTGGTGTTTCAGTTGGTGTATTAACATCT
CTTCTCATCAATTATCTCAGAAATTAAAAAGCTGAGTGCAAACAATAAAGACAATCTTTG
GCTATTACAGAATCCGAATAACCATATGAAGATGATGATGCTAAGAGAAGAAAAGTTATC
TAAGAGCTCATTAAAACTAACAAAGGCAAGTACAAAGAATTCCATAGACTTAAAATTAAG
GATGGTGCATTAGTTGGAAATTCATTACTTGATGAAATCAGATTCCTTGACAGTGCTCTT
TGTGAAATAGAGAAAGTTCCTAACCTCATTATTGATTTAGGATTAGACATCTTTGAAGAG
GTAGTTAACCCTATTGCAAGACACACTCATAAAGATACTTTTATTAATACAAACTCATTT
ATTTTATCCTATGCTGCATTTGGTCCTTCTTAAATGATTCAAAATGCTAAGAGATGGTTA
AGAGCTGGTCCTCGTTATCACAATTACTTTAGACCTGACTAATCAAAGGTTTTGATTGTG
ACATAAGATGGCCTTTGCCCTGGTTTGAATGTCGTCATTAGAGAAATTGTTATGCACTTA
AATTACAACTATTAGGTTGAATAAATCTATGGAGCCAAATTTGGTTGGAAAGGTATTTAC
GAGGAAGACTGGGTTCAATTAATTCCCAGCTAAATTAAAACTATCCATCACAGAGGTGGT
ACCTTCATAGGAACTTCAAGAACTGGTTTTGATAGAGATAAAATTATTGAAACACTTATC
AAGCACAATTTTAATTAGCTCTATGTCATCTGCGGAAACAAATCCATGGGTCAAGTTCTT
GAACTTTATTATGAAATCAGAAGAAGAAAACTCAATATTGGTGTTTGTAACATTCCAAGA
AGCGTCGATAATGATATTCCTATTATCGATGAATCATTTGGTTATTAATCTTGTGTTGAA
GAAGCTTAAAGAGTAATTGATAGTGCTTGGGTCGAATCTCGTTCCCATAGAAATGGTGTT
GGTATTATTAGAGTTATGGGATAAAAGAGTGGATTTATTGCTATGGGTGCTTGTCTTGCA
TCTCGTTGTGTAAATGTATGTTTGATCCCTGAATTTAATTACGATCTATATGGAAAGAGT
GGTGTCCTTGACTATATCAAATAGAGAATTATTAAGAAGGGTCATTGTATAGTTGTTGTT
TCTGAAGGAGCCAACAGCTCATTTGCTGATGCTGATGATGTTCCTGGTGATGTTGGTAAA
TTCTTATAAGATGAAATTCAAAAGTTTTGTGCTTAAGACAATACTGAAGTTACTGTAAAA
TATTTTGATCCTTTATATATGCTTCGTGCTGTTAGAGCTAATGCTTATGACACCAAATAG
TGCAGTACTCTTGCTTAAAACGCAGTACATGGTTTAATGGCAGGTTTCACTGGGTTTTCT
ATTGGTCACGTCCATAACAAGACTTGTTTCATACCTCTTGAAGAAATGCTTAGTGGAAAT
TATAGAAATAGAATAGTAGCAAGTAACAAAAACTGGTAGAGATTACTAGCTTCTACTGGT
TAGCCATCCTTCATCAATGAAGAAGAATTAAAGAAAATAATTGGAACTAGCAAATAAGAA
TGA>TTHERM_00338460(gene)
ATTTACAATACAAACTATACAGAAAATATATATTTAAAATATAATGAAAATTTTAGGTTT
GCATAATTAATTAATGAATTGATTGATTGATTCGCTTAGCTATTCAATAAAAACATGAAA
TAAATCAGTGATATATTTTTTTATTGATTGAATGATGCTTGCTACTTTAAAATGGATTAG
AGAGTATAGTGTCAATATCTTTTATTTTAGTAAATTAAAAAAATAAAAATATTAACAAGA
ATCTTAATAGCGATCTTCTAAAAATTGTTTGCAAATTTATTGATAACAAATAGTAATACA
ATAAAATATATATTGAAAAAAAGCTAAAAAGGATTTGATTTTTAATAACAATTTAAAAGA
ATATTAAAGAGTAGAAATAGAAATGGAAAAAAACACTGTTTTGACTACAAGCATTGGTGT
TTCAGTTGGTGTATTAACATCTCTTCTCATCAATTATCTCAGAAATTAAAAAGCTGAGTG
CAAACAATAAAGACAATCTTTGGCTATTACAGAATCCGAATAACCATATGAAGATGATGA
TGCTAAGAGAAGAAAAGTTATCTAAGAGCTCATTAAAACTAACAAAGGCAAGTACAAAGA
ATTCCATAGACTTAAAATTAAGGATGGTGCATTAGTTGGAAATTCATTACTTGATGAAAT
CAGATTCCTTGACAGTGCTCTTTGTGAAATAGAGAAAGTTCCTAACCTCATTATTGATTT
AGGATTAGACATCTTTGAAGAGGTAGTTAACCCTATTGCAAGACACACTCATAAAGATAC
TTTTATTAATACAAACTCATTTATTTTATCCTATGCTGCATTTGGTCCTTCTTAAATGAT
TCAAAATGCTAAGAGATGGTTAAGAGCTGGTCCTCGTTATCACAATTACTTTAGACCTGA
CTAATCAAAGGTTTTGATTGTGACATAAGATGGCCTTTGCCCTGGTTTGAATGTCGTCAT
TAGAGAAATTGTTATGCACTTAAATTACAACTATTAGGTTGAATAAATCTATGGAGCCAA
ATTTGGTTGGAAAGGTATTTACGAGGAAGACTGGGTTCAATTAATTCCCAGCTAAATTAA
AACTATCCATCACAGAGGTGGTACCTTCATAGGAACTTCAAGAACTGGTTTTGATAGAGA
TAAAATTATTGAAACACTTATCAAGCACAATTTTAATTAGCTCTATGTCATCTGCGGAAA
CAAATCCATGGGTCAAGTTCTTGAACTTTATTATGAAATCAGAAGAAGAAAACTCAATAT
TGGTGTTTGTAACATTCCAAGAAGCGTCGATAATGATATTCCTATTATCGATGAATCATT
TGGTTATTAATCTTGTGTTGAAGAAGCTTAAAGAGTAATTGATAGTGCTTGGGTCGAATC
TCGTTCCCATAGAAATGGTGTTGGTATTATTAGAGTTATGGGATAAAAGAGTGGATTTAT
TGCTATGGGTGCTTGTCTTGCATCTCGTTGTGTAAATGTATGTTTGATCCCTGAATTTAA
TTACGATCTATATGGAAAGAGTGGTGTCCTTGACTATATCAAATAGAGAATTATTAAGAA
GGGTCATTGTATAGTTGTTGTTTCTGAAGGAGCCAACAGCTCATTTGCTGATGCTGATGA
TGTTCCTGGTGATGTTGGTAAATTCTTATAAGATGAAATTCAAAAGTTTTGTGCTTAAGA
CAATACTGAAGTTACTGTAAAATATTTTGATCCTTTATATATGCTTCGTGCTGTTAGAGC
TAATGCTTATGACACCAAATAGTGCAGTACTCTTGCTTAAAACGCAGTACATGGTTTAAT
GGCAGGTTTCACTGGGTTTTCTATTGGTCACGTCCATAACAAGACTTGTTTCATACCTCT
TGAAGAAATGCTTAGTGGAAATTATAGAAATAGAATAGTAGCAAGTAACAAAAACTGGTA
GAGATTACTAGCTTCTACTGGTTAGCCATCCTTCATCAATGAAGAAGAATTAAAGAAAAT
AATTGGAACTAGCAAATAAGAATGAGAATTAAAATAATAAATTGAATAATTATAATATTT
TAAAAGTTTTCTACTTTGAATACAAAAAGTAGTTTGTGTAGATTTTAAGAAATATTTTCT
TTAAAAATGAAGTTTTCG>TTHERM_00338460(protein)
MEKNTVLTTSIGVSVGVLTSLLINYLRNQKAECKQQRQSLAITESEQPYEDDDAKRRKVI
QELIKTNKGKYKEFHRLKIKDGALVGNSLLDEIRFLDSALCEIEKVPNLIIDLGLDIFEE
VVNPIARHTHKDTFINTNSFILSYAAFGPSQMIQNAKRWLRAGPRYHNYFRPDQSKVLIV
TQDGLCPGLNVVIREIVMHLNYNYQVEQIYGAKFGWKGIYEEDWVQLIPSQIKTIHHRGG
TFIGTSRTGFDRDKIIETLIKHNFNQLYVICGNKSMGQVLELYYEIRRRKLNIGVCNIPR
SVDNDIPIIDESFGYQSCVEEAQRVIDSAWVESRSHRNGVGIIRVMGQKSGFIAMGACLA
SRCVNVCLIPEFNYDLYGKSGVLDYIKQRIIKKGHCIVVVSEGANSSFADADDVPGDVGK
FLQDEIQKFCAQDNTEVTVKYFDPLYMLRAVRANAYDTKQCSTLAQNAVHGLMAGFTGFS
IGHVHNKTCFIPLEEMLSGNYRNRIVASNKNWQRLLASTGQPSFINEEELKKIIGTSKQE