Gene Model Identifier
TTHERM_00339640
Standard Name
VMA1
(Vacuolar Membrane Atpase)
Aliases
PreTt06105 | 34.m00342 | 3689.m00136
Description
VMA1 vacuolar H+-ATPase A subunit; V-type ATPase, A subunit family protein; V-type ATP synthase catalytic alpha chain
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
>TTHERM_00339640(coding)
ATGGCTTAAAGAAAAACTGCTTTAGAAGAAATTGCCGAAAAGGAATCCAACTTCGGTAAA
ATTTTCAAGGTCGCCGGTCCTCTGGTCGTTGCTGAACAAATGGTTGGTGCTAAGATGTAC
GAATTGGTTAAAGTCGGTTGGAATAAATTGGTCGGTGAAATTATCAAGCTCGATGGTGAC
ACTGCATCTATTCAATGTTACGAAGATACTTATGGTTTGACTGTTGGTGATCCAGTCATG
AGAACTAAAGAACCTCTTTCAGTCGAACTTGGTCCTGGTATTTTGAATGGTATTTTTGAC
GGTATCTAAAGACCTCTTGGTTCTATTGCTTAGGTTTCAGGTTCAGCTTTCATCCCTAGA
GGTGTTGATGTCCCTATTTTAGATGGAGATAAGTAATGGCATTTTATTCCTAGACCTGAT
ATTAAGGTTGGCTCTAAGCTTACTGGTGGTGATATCTTCGGTCACTGTCACGAAAACAAC
TTGTTCTGTAAGCACAACATTATGCTTCCTCCCAAGGGTAAGGGTCACGTCACTTACATT
GCTCCTGAAGGCTAATACACCATCCATGAAAAAGTTTTAGAACTCGAAATTGATGGCAAG
AAGACTTCTTACAAAATGTCTCACTTCTGGCCTGTCAGATAAGCTCGTCCCGTTTTAGAA
AGACTCTAAGGTAATGTTCCTCTTCTTACTGGTCAAAGAGTTTTGGATGCTCTTTTCCCT
TCAGTCTTGGGTGGTACTTGCGCTATTCCTGGTGCTTTTGGTTGCGGTAAAACTTGTATT
TCTTAAGCCTTATCTAAATACTCTAACTCCGAAGTCATCGTTTACGTCGGTTGTGGTGAA
AGAGGTAACGAAATGGCTGAAGTACTTACTGATTTCCCTGAATTGAAAACTGAAATTAAT
GGTAAGAAAGAATCTATTATGCAACGTACTTGTTTGGTCGCTAATACATCTAACATGCCT
GTCGCTGCCAGAGAAGCTTCTATTTACACTGGTATTACTCTTGCTGAATACTTCAGAGAT
ATGGGTTACAACGTTAGTATGATGGCTGACTCAACTTCTCGTTGGGCTGAAGCCTTGAGA
GAAATTTCTGGTCGTCTTGCTTAGATGCCTGCTGAATAAGGTTATCCTGCTTATCTTGCC
AATAAGCTTGCCTTCTTCTACGAAAGAGCTGGTCGTGTCAAATGTTAGGGTTCTCCCAAC
AGAGAAGGTTCTATTACAATTGTCGGTGCCGTCTCTCCTCCTGGTGGTGATTTCACTGAT
CCAGTTACAGTCTCTACTCTTGCTATCGTACAAGTTTTCTGGGGTCTCGATAAGAAATTA
GCTCAACGTAAACATTTCCCTTCAGTCGACTGGAACAAATCTACATCCAACTACGAAAGA
CAATTAGATCCTTACTTCAACAACTTCGATCCTGATTTCTCTAATTTACGTACTTCTATT
AAAAAAATCTTATAAGAAGAAAATGAATTGAATGAAATCGTCCAATTGGTCGGTAAGGAC
TCTCTTTCTGAAGATTAAAAGCTTACCTTAGAAGTCGCTAGAGTTATTAGAGAAGATTTC
TTACAATAAAACGCTTTCTCAGATTATGATTACATGTGTCCCTTAGAAAAGACTATTGGT
ATGATGAGAGGTATTTTCACTTACTACGACAATGCTAGAAGAACCATTGTTGAATCTTCT
GGTGAAACCAAGATTAACTGGAACGAAATTCTCTTCCAAACCAAGCCTCTCTTCCTCTAA
CTTACTGATATGAAGAGACTTGACCCTAAAAAACCTAAATAAGAAGTCAATCAATATTTC
AGTAAATACGTTGAAGATGTCAATCTTGCTTTCAGAAAAATCAATGACCGTTGA
>TTHERM_00339640(gene)
AATTTTCTTTGGGAGAGAGGAAGATATATTAACAATTTCTTTTATTCTTTGATATCAATA
ATTTGTGAGATTTATTTGATCATACATTTACATATGAATTTTGATTTCTATCAAACAATA
TTGAATACTAAAAAATTAATTAAAAGGGTAGGTAGGTGATTGATAAACAGCTAAATTAAT
AATAAAAAGATTAAAATGAAATGAAGTTCGCTTGCTTGTATATAAATATTTTTATGCATA
TTTATCGTAGAATAGATATGGAAGTATAAACATTATTGATAGATAGAAAGAAAGAATGAA
ACAAGCCATAAATCAATCAATCAATCAATTAATTAATAAATCATTAATGAAGATAAGTAA
GTATTAGAGATAGGAAGAAAGATTAATCAATAATAAAAGATAATAAAGCGAAATAAAAAA
ATATAAAAGGGGAAAACAAATAAAATAAAAATAAAAATAAACAATTAAAGAAAACAAAAA
TTAATATAATAATAAAAAATAAAATAAAAACTAAGTAAGAAAGAAGTCTAATTCACTCAA
ATGGCTTAAAGAAAAACTGCTTTAGAAGAAATTGCCGAAAAGGAATCCAACTTCGGTAAA
ATTTTCAAGGTCGCCGGTCCTCTGGTCGTTGCTGAACAAATGGTTGGTGCTAAGATGTAC
GAATTGGTTAAAGTCGGTTGGAATAAATTGGTCGGTGAAATTATCAAGCTCGATGGTGAC
ACTGCATCTATTCAATGTTACGAAGATACTTATGGTTTGACTGTTGGTGATCCAGTCATG
AGAACTAAAGAACCTCTTTCAGTCGAACTTGGTCCTGGTATTTTGAATGGTATTTTTGAC
GGTATCTAAAGACCTCTTGGTTCTATTGCTTAGGTTTCAGGTTCAGCTTTCATCCCTAGA
GGTGTTGATGTCCCTATTTTAGATGGAGATAAGTAATGGCATTTTATTCCTAGACCTGAT
ATTAAGGTTGGCTCTAAGCTTACTGGTGGTGATATCTTCGGTCACTGTCACGAAAACAAC
TTGTTCTGTAAGCACAACATTATGCTTCCTCCCAAGGGTAAGGGTCACGTCACTTACATT
GCTCCTGAAGGCTAATACACCATCCATGAAAAAGTTTTAGAACTCGAAATTGATGGCAAG
AAGACTTCTTACAAAATGTCTCACTTCTGGCCTGTCAGATAAGCTCGTCCCGTTTTAGAA
AGACTCTAAGGTAATGTTCCTCTTCTTACTGGTCAAAGAGTTTTGGATGCTCTTTTCCCT
TCAGTCTTGGGTGGTACTTGCGCTATTCCTGGTGCTTTTGGTTGCGGTAAAACTTGTATT
TCTTAAGCCTTATCTAAATACTCTAACTCCGAAGTCATCGTTTACGTCGGTTGTGGTGAA
AGAGGTAACGAAATGGCTGAAGTACTTACTGATTTCCCTGAATTGAAAACTGAAATTAAT
GGTAAGAAAGAATCTATTATGCAACGTACTTGTTTGGTCGCTAATACATCTAACATGCCT
GTCGCTGCCAGAGAAGCTTCTATTTACACTGGTATTACTCTTGCTGAATACTTCAGAGAT
ATGGGTTACAACGTTAGTATGATGGCTGACTCAACTTCTCGTTGGGCTGAAGCCTTGAGA
GAAATTTCTGGTCGTCTTGCTTAGATGCCTGCTGAATAAGGTTATCCTGCTTATCTTGCC
AATAAGCTTGCCTTCTTCTACGAAAGAGCTGGTCGTGTCAAATGTTAGGGTTCTCCCAAC
AGAGAAGGTTCTATTACAATTGTCGGTGCCGTCTCTCCTCCTGGTGGTGATTTCACTGAT
CCAGTTACAGTCTCTACTCTTGCTATCGTACAAGTTTTCTGGGGTCTCGATAAGAAATTA
GCTCAACGTAAACATTTCCCTTCAGTCGACTGGAACAAATCTACATCCAACTACGAAAGA
CAATTAGATCCTTACTTCAACAACTTCGATCCTGATTTCTCTAATTTACGTACTTCTATT
AAAAAAATCTTATAAGAAGAAAATGAATTGAATGAAATCGTCCAATTGGTCGGTAAGGAC
TCTCTTTCTGAAGATTAAAAGCTTACCTTAGAAGTCGCTAGAGTTATTAGAGAAGATTTC
TTACAATAAAACGCTTTCTCAGATTATGATTACATGTGTCCCTTAGAAAAGACTATTGGT
ATGATGAGAGGTATTTTCACTTACTACGACAATGCTAGAAGAACCATTGTTGAATCTTCT
GGTGAAACCAAGATTAACTGGAACGAAATTCTCTTCCAAACCAAGCCTCTCTTCCTCTAA
CTTACTGATATGAAGAGACTTGACCCTAAAAAACCTAAATAAGAAGTCAATCAATATTTC
AGTAAATACGTTGAAGATGTCAATCTTGCTTTCAGAAAAATCAATGACCGTTGATTTATC
TGATATACTATAATTTTATATATTGCTACCCTATACTTATTTTGATATACCTCTGATATC
TTCATTCATTTGTACTAATCATAATATATCAAATAAATTATATTAATTTCAGTATTTCTA
TGGTTAATATAGTGTATTTTGTTTTTTTTAAAGGATATATTCTTCTCTTTATAAGT
>TTHERM_00339640(protein)
MAQRKTALEEIAEKESNFGKIFKVAGPLVVAEQMVGAKMYELVKVGWNKLVGEIIKLDGD
TASIQCYEDTYGLTVGDPVMRTKEPLSVELGPGILNGIFDGIQRPLGSIAQVSGSAFIPR
GVDVPILDGDKQWHFIPRPDIKVGSKLTGGDIFGHCHENNLFCKHNIMLPPKGKGHVTYI
APEGQYTIHEKVLELEIDGKKTSYKMSHFWPVRQARPVLERLQGNVPLLTGQRVLDALFP
SVLGGTCAIPGAFGCGKTCISQALSKYSNSEVIVYVGCGERGNEMAEVLTDFPELKTEIN
GKKESIMQRTCLVANTSNMPVAAREASIYTGITLAEYFRDMGYNVSMMADSTSRWAEALR
EISGRLAQMPAEQGYPAYLANKLAFFYERAGRVKCQGSPNREGSITIVGAVSPPGGDFTD
PVTVSTLAIVQVFWGLDKKLAQRKHFPSVDWNKSTSNYERQLDPYFNNFDPDFSNLRTSI
KKILQEENELNEIVQLVGKDSLSEDQKLTLEVARVIREDFLQQNAFSDYDYMCPLEKTIG
MMRGIFTYYDNARRTIVESSGETKINWNEILFQTKPLFLQLTDMKRLDPKKPKQEVNQYF
SKYVEDVNLAFRKINDR
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
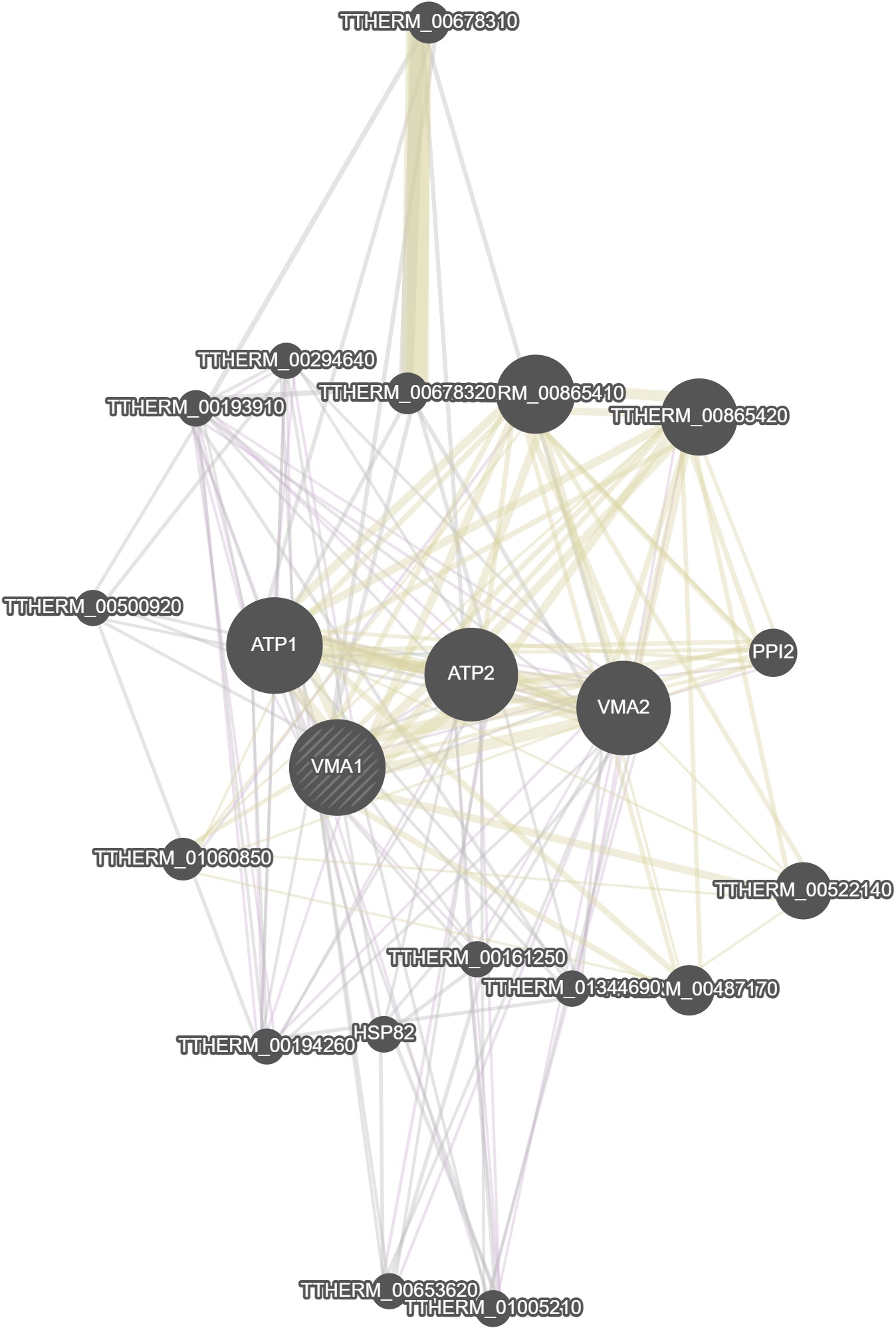
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences