Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00348660Standard Name
CTH46 (CaTHepsin 46)Aliases
PreTt05278 | 36.m00242 | 3703.m00104Description
CTH46 papain family cysteine protease; hypothetical proteinGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- obsolete integral component of mitochondrial membrane (IEA) | GO:0032592
Molecular Function
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- ABC-type maltose transporter activity (IEA) | GO:0015423
- ABC-type oligosaccharide transporter activity (IEA) | GO:0015422
- alpha-humulene synthase activity (IEA) | GO:0080017
- Cys-tRNA(Pro) hydrolase activity (IEA) | GO:0043907
- cytoskeletal anchor activity (IEA) | GO:0008093
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- insulin-like growth factor II binding (IEA) | GO:0031995
- interleukin-10 receptor binding (IEA) | GO:0005141
- NAD+ synthase activity (IEA) | GO:0008795
- obsolete D-lysine oxidase activity (IEA) | GO:0043912
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- tumor necrosis factor receptor binding (IEA) | GO:0005164
Biological Process
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- auditory receptor cell fate commitment (IEA) | GO:0009912
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- embryonic brain development (IEA) | GO:1990403
- folic acid-containing compound metabolic process (IEA) | GO:0006760
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- laminaritriose transport (IEA) | GO:2001097
- maintenance of protein location in peroxisome (IEA) | GO:0072664
- negative regulation by organism of cell-mediated immune response of other organism involved in symbiotic interaction (IEA) | GO:0052278
- negative regulation of chemokine-mediated signaling pathway (IEA) | GO:0070100
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of glycine secretion, neurotransmission (IEA) | GO:1904625
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete activation of MAPK activity (IEA) | GO:0000187
- obsolete bent DNA binding (IEA) | GO:0006326
- obsolete negative regulation by organism of defense-related symbiont ethylene-mediated signal transduction pathway (IEA) | GO:0052479
- obsolete negative regulation of histone H2A-H2B dimer displacement (IEA) | GO:1902650
- obsolete positive regulation by symbiont of host phytoalexin production (IEA) | GO:0052344
- obsolete regulation of toxin transport (IEA) | GO:1902007
- obsolete satellite DNA binding (IEA) | GO:0006329
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- photosynthetic NADP+ reduction (IEA) | GO:0009780
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of fatty acid beta-oxidation (IEA) | GO:0032000
- positive regulation of flower development (IEA) | GO:0009911
- positive regulation of peroxisome proliferator activated receptor signaling pathway (IEA) | GO:0035360
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of stress response to copper ion (IEA) | GO:1903855
- protein localization to old growing cell tip (IEA) | GO:1903858
- protein localization to vacuole (IEA) | GO:0072665
- pteridine biosynthetic process (IEA) | GO:0006728
- regulation of (1->3)-beta-D-glucan biosynthetic process (IEA) | GO:0032953
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of rhamnose catabolic process (IEA) | GO:0043463
- tetrasaccharide transport (IEA) | GO:2001098
- vestibular receptor cell fate commitment (IEA) | GO:0060115
- vitamin A metabolic process (IEA) | GO:0006776
- xanthophyll biosynthetic process (IEA) | GO:0016123
- xanthophyll catabolic process (IEA) | GO:0016124
 Domains
Domains
No Data fetched for Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
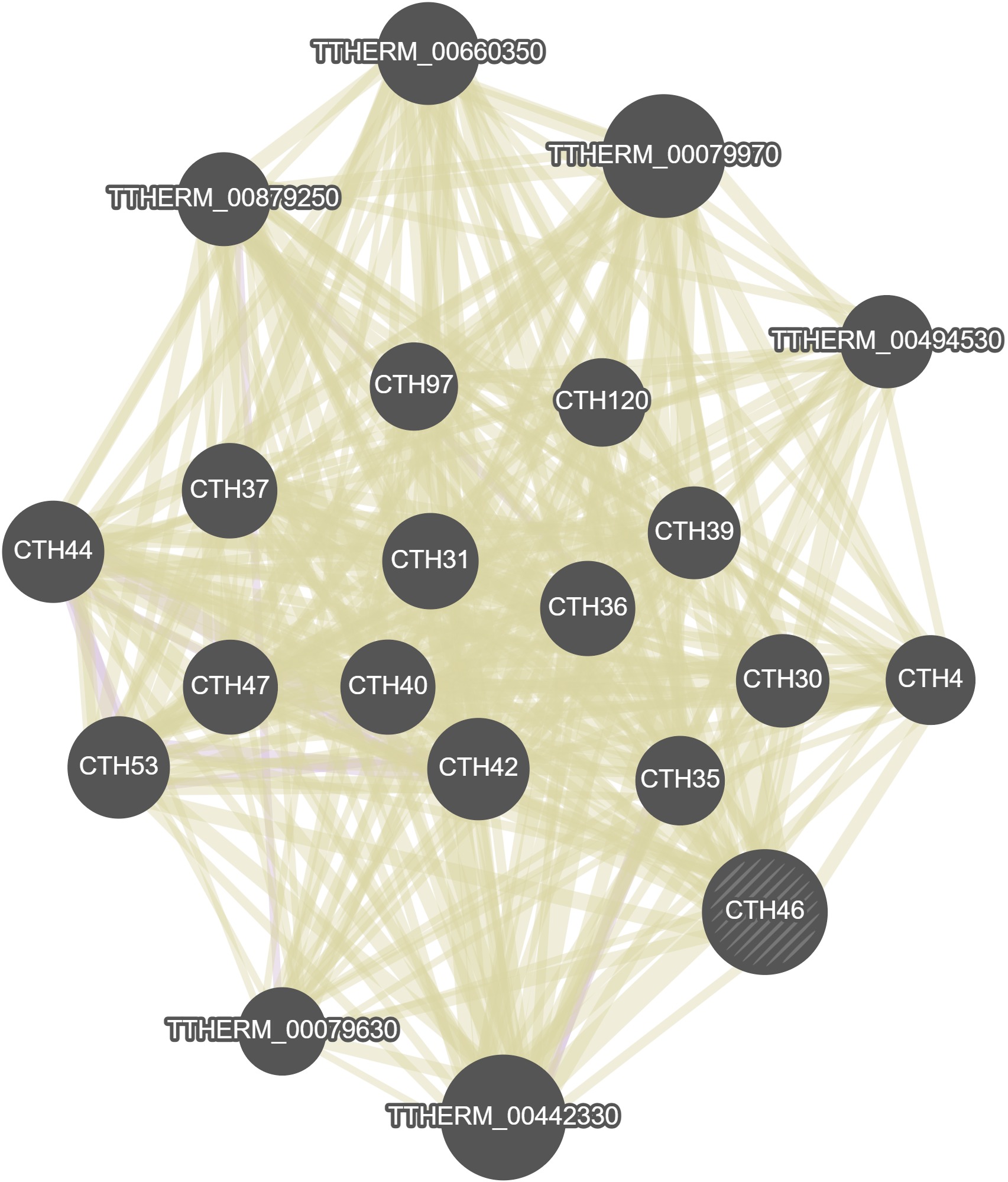
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00348660(coding)
ATGAAGAATTAAAAATAAACAAGCTAATAAAGCGAAGCAGCTAAGCAAGCTTAGTTAAAC
CGCTAAAAGAAAAAAGGAAGTTCACTAGGATTTTTTAGCAAGTATTCAACTTAAATTAGT
GTTGCTTTGTTTGGTTCCCTTTTGTTAGGTGCAATTATTTCAGTAATGATACCAACTGGC
AAGAAATTAACTGAAAATGTTATTGTTGAAGAAGAAATAGCTTAACACAATGCATAAAAT
TATTCTTTTAAAATAGGTCCTAATACAATCTTTAAGGACACTACTCTTGCTTCAACTTAA
AAATTGTTTTTGAATTAAATTTCACATAAATAATCTTTAATAAGATGTCCTTCTGAATAG
GATTAAAATATAATAGATTTGTCCTTTAATTTCCATACTAAATATCCCTAGTGCGTCCGT
CCAATTGCAAATTAAGGAAAAGATTGCAGTGCTTCTTATTCAATAGCCGCTGTTTCTTCT
GTTGCTGATAGACTCTGTATGGCATCTGAGGGAGATTTTAACTTCGGTCTTAGTGCTTAA
CCTACCATCTCTTGTTATGAAAATTAATCATACAAGTGTGAAGGTGGTTATGTATCTAAA
ACTTTCTAAAAAGGAAAGACCACTGGTTTTGTCAAAGAAGAATGTTTACCTTATCATGGT
ACTGATTCAAATGAAGGATGCTCACTTATTGATAAATGTGAACATTTCAAGATTTATGAT
TATTGTGTTTCAGCAGGTTAAGAAAGCATTAAACGTGAAATCATGTTAAATGGACCTGTT
GTATCATTAATGAACGTCTTTTCTGATTTCTTAGTTTACAAGTCAGGTGTTTACCGTGTT
TTAGAAAATGCAGCAAAACTAAAAGGTCAATAAGCAGTTAAAATTATTGGTTGGGATATT
GATCCTCTTACTAAGGACTACTATTGGATAATAGAAAACTCTTGGGGAGAAGAATGGGGT
TTAAACGGTTTAGCTTACGTAGCTATGGGATAAGAAGAATTAAGATTAGAAGAATATGCT
CTTGCTGCAATTACCCTATAACAAGCCGAAACAGCTTCATCCTAATAAGCAAAAGCCTAA
AGTCAAGGATCTACTCTCAATTATGACGAAATTTGA>TTHERM_00348660(gene)
TAAAGATACACATCATAAATATTTTTATAAATAATAAATATATTAAAAAATAAATTAATC
TGTTATTTGCAAGATAAATATGAAGAATTAAAAATAAACAAGCTAATAAAGCGAAGCAGC
TAAGCAAGCTTAGTTAAACCGCTAAAAGAAAAAAGGAAGTTCACTAGGATTTTTTAGCAA
GTATTCAACTTAAATTAGTGTTGCTTTGTTTGGTTCCCTTTTGTTAGGTGCAATTATTTC
AGTAATGATACCAACTGGCAAGAAATTAACTGAAAATGTTATTGTTGAAGAAGAAATAGC
TTAACACAATGCATAAAATTATTCTTTTAAAATAGGTCCTAATACAATCTTTAAGGACAC
TACTCTTGCTTCAACTTAAAAATTGTTTTTGAATTAAATTTCACATAAATAATCTTTAAT
AAGATGTCCTTCTGAATAGGATTAAAATATAATAGATTTGTCCTTTAATTTCCATACTAA
ATATCCCTAGTGCGTCCGTCCAATTGCAAATTAAGGAAAAGATTGCAGTGCTTCTTATTC
AATAGCCGCTGTTTCTTCTGTTGCTGATAGACTCTGTATGGCATCTGAGGGAGATTTTAA
CTTCGGTCTTAGTGCTTAACCTACCATCTCTTGTTATGAAAATTAATCATACAAGTGTGA
AGGTGGTTATGTATCTAAAACTTTCTAAAAAGGAAAGACCACTGGTTTTGTCAAAGAAGA
ATGTTTACCTTATCATGGTACTGATTCAAATGAAGGATGCTCACTTATTGATAAATGTGA
ACATTTCAAGATTTATGATTATTGTGTTTCAGCAGGTTAAGAAAGCATTAAACGTGAAAT
CATGTTAAATGGACCTGTTGTATCATTAATGAACGTCTTTTCTGATTTCTTAGTTTACAA
GTCAGGTGTTTACCGTGTTTTAGAAAATGCAGCAAAACTAAAAGGTCAATAAGCAGTTAA
AATTATTGGTTGGGATATTGATCCTCTTACTAAGGACTACTATTGGATAATAGAAAACTC
TTGGGGAGAAGAATGGGGTTTAAACGGTTTAGCTTACGTAGCTATGGGATAAGAAGAATT
AAGATTAGAAGAATATGCTCTTGCTGCAATTACCCTATAACAAGCCGAAACAGCTTCATC
CTAATAAGCAAAAGCCTAAAGTCAAGGATCTACTCTCAATTATGACGAAATTTGATTTGA
GAGCAAATAAGAAAACGATAAGTTATCTAAATTTATAGAAATTTGGTATTTATAGCACAT
TTTTCAAACATTTTTGTATTTATTTTAGCTTAAATATAATGAACTATTGATTCTTATTTT
GTGGGTTAATGATAAAATAATAAATTTGTGAAAATTTTAATGAAATAAATTCTAAAAACT
TTAATTATTTAACATAGAATTATTAAAATCTATGAGTTTCATAAAAATTTA>TTHERM_00348660(protein)
MKNQKQTSQQSEAAKQAQLNRQKKKGSSLGFFSKYSTQISVALFGSLLLGAIISVMIPTG
KKLTENVIVEEEIAQHNAQNYSFKIGPNTIFKDTTLASTQKLFLNQISHKQSLIRCPSEQ
DQNIIDLSFNFHTKYPQCVRPIANQGKDCSASYSIAAVSSVADRLCMASEGDFNFGLSAQ
PTISCYENQSYKCEGGYVSKTFQKGKTTGFVKEECLPYHGTDSNEGCSLIDKCEHFKIYD
YCVSAGQESIKREIMLNGPVVSLMNVFSDFLVYKSGVYRVLENAAKLKGQQAVKIIGWDI
DPLTKDYYWIIENSWGEEWGLNGLAYVAMGQEELRLEEYALAAITLQQAETASSQQAKAQ
SQGSTLNYDEI