Gene Model Identifier
TTHERM_00355040
Standard Name
SAP1
(Swc4-associated protein 1)
Aliases
PreTt27490 | 37.m00207 | 3836.m02293
Description
SAP1 Member of the SWR complex; Zinc finger C2H2-type
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
>TTHERM_00355040(coding)
ATGAATAATTAGCAAGTGGATTAAGGAATGTAAGATGGTTAAAACGGTTAATCTGAAAAG
AAAACTAGAAGAACCCGTAAAGATTAAAATGACAGAAACTATAATTGCGGTTGTGGTAAA
TCATATTTATCATACCCTGCTTTATATACTCATCTTAAACAAAAACACGATGGTAAACCA
CCTGAGGGTACTAGTCTTCCAGGACCATAAGGCAAAGCAAATAGCAGAGGACGTCCTCCC
AAAAAAGAAGATGGTGATCAAAATAATAAAGAAGAAGAGAAGAGCGTTGGATCTAACGAA
GCAGTTGATGTGCTTGAAGAAATATTGAACTTTTTAGATACTATTGATAAGAATTATAGA
AGAAATAAAAATGACGAGTGGACTGAAGATAATAAGCTAAGCACTATATTCCCTATTGAA
TTCTTCCAAGGTAAGGAAGAAACTGAGTATAAAGCAATTTTAGATGGGATTAAGCAACTT
GAAGATGATCCAGCAAACTATATTTATGAAGAAATAGATCCAAAAGAGGAAATCAAGAAG
ACTGAAATGAATAGAATATTTGTTTTATTCTTAAATTACATAGCTAAAGGTGTATAAAAC
GAAGCATTAAAAGAGATTATGATATTCCTCTGTTTTTATCGTAAAGCATTGAATTAATAT
GGCTGGGATGCTCTTGAATAAAAAATAAACAATGAATTGCAATAATAAAATATGCTTGTT
AATGGCTAAAACGGAGATAATCCAGAATTAAATGGAAACTCTGAAGCAGCACCACCCTAA
TAGCAATATTAAATTGAATAATCTAGAAGACTATAAGAGTACTGCCAAATAAATAATGGT
GACGATTCTTTCTTAATATGCAACGATTTAGTCATCGAAGTCTTACCTAATTATTTCAGA
GAATATCCCAATTCAAATGATTTAGTTATTATAGGTCCATCTGATGAATAACTTAAAAAT
GCAGTCTATATAATTTAACATTTTGCTAACTGGCTTTTTGCAAACAGATATACAAATACA
AAGTTGGTTATTAAAGCAGATGATGAATGA
>TTHERM_00355040(gene)
CTAAATTAATAATAATTAATTAATAATTATATACATTCTAAAGATATTTTTTAGTGATTC
AGAAAAGAATTTATCTAATAAAAGTAGAAAGCGTATTTTATAAATTTAGCACCTACAAAT
ATTAAGCATTTCATAGTATAGTAAAATGAATAATTAGCAAGTGGATTAAGGAATGTAAGA
TGGTTAAAACGGTTAATCTGAAAAGAAAACTAGAAGAACCCGTAAAGATTAAAATGACAG
AAACTATAATTGCGGTTGTGGTAAATCATATTTATCATACCCTGCTTTATATACTCATCT
TAAACAAAAACACGATGGTAAACCACCTGAGGGTACTAGTCTTCCAGGACCATAAGGCAA
AGCAAATAGCAGAGGACGTCCTCCCAAAAAAGAAGATGGTGATCAAAATAATAAAGAAGA
AGAGAAGAGCGTTGGATCTAACGAAGCAGTTGATGTGCTTGAAGAAATATTGAACTTTTT
AGATACTATTGATAAGAATTATAGAAGAAATAAAAATGACGAGTGGACTGAAGATAATAA
GCTAAGCACTATATTCCCTATTGAATTCTTCCAAGGTAAGGAAGAAACTGAGTATAAAGC
AATTTTAGATGGGATTAAGCAACTTGAAGATGATCCAGCAAACTATATTTATGAAGAAAT
AGATCCAAAAGAGGAAATCAAGAAGACTGAAATGAATAGAATATTTGTTTTATTCTTAAA
TTACATAGCTAAAGGTGTATAAAACGAAGCATTAAAAGAGATTATGATATTCCTCTGTTT
TTATCGTAAAGCATTGAATTAATATGGCTGGGATGCTCTTGAATAAAAAATAAACAATGA
ATTGCAATAATAAAATATGCTTGTTAATGGCTAAAACGGAGATAATCCAGAATTAAATGG
AAACTCTGAAGCAGCACCACCCTAATAGCAATATTAAATTGAATAATCTAGAAGACTATA
AGAGTACTGCCAAATAAATAATGGTGACGATTCTTTCTTAATATGCAACGATTTAGTCAT
CGAAGTCTTACCTAATTATTTCAGAGAATATCCCAATTCAAATGATTTAGTTATTATAGG
TCCATCTGATGAATAACTTAAAAATGCAGTCTATATAATTTAACATTTTGCTAACTGGCT
TTTTGCAAACAGATATACAAATACAAAGTTGGTTATTAAAGCAGATGATGAATGAAAGAG
CGTGCAGTCAGTTGTATAAGTAATTTACATTCTAACTGCATATATGTAAATTCTAAAAAA
AATAATATTACATTGCATATAAAAAAATCTATTCTTTTAAGACACATATTATTCTCTAAA
GCAGATATTAAAGTAATAGTTAATGATAGTTCTCAAAATTAGCAAACAGAATAAAAAACT
TAAAGCATAAAAAATCAATTCAATACCACATTTTTACAAAATTTAATTTATTACACATAT
TCACTCCAATTTCAAGAATGAAGTAAATAATTATTGTTTTTTGCTATTTTTATCAGTGGG
TTGATTAATTGATAATATAATAACCAGCTTACTTATCTAACTAAGCTTACTTTATTTGCT
CATTTATTAACTAACCTAAATATCAATCATATTTTAAACTTTTATATTACATAAGTTATT
TTTATATTTGTAAATCTTAAGATATTTAGTTAACAAATTCAAATAAACTAAATAAAC
>TTHERM_00355040(protein)
MNNQQVDQGMQDGQNGQSEKKTRRTRKDQNDRNYNCGCGKSYLSYPALYTHLKQKHDGKP
PEGTSLPGPQGKANSRGRPPKKEDGDQNNKEEEKSVGSNEAVDVLEEILNFLDTIDKNYR
RNKNDEWTEDNKLSTIFPIEFFQGKEETEYKAILDGIKQLEDDPANYIYEEIDPKEEIKK
TEMNRIFVLFLNYIAKGVQNEALKEIMIFLCFYRKALNQYGWDALEQKINNELQQQNMLV
NGQNGDNPELNGNSEAAPPQQQYQIEQSRRLQEYCQINNGDDSFLICNDLVIEVLPNYFR
EYPNSNDLVIIGPSDEQLKNAVYIIQHFANWLFANRYTNTKLVIKADDE
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
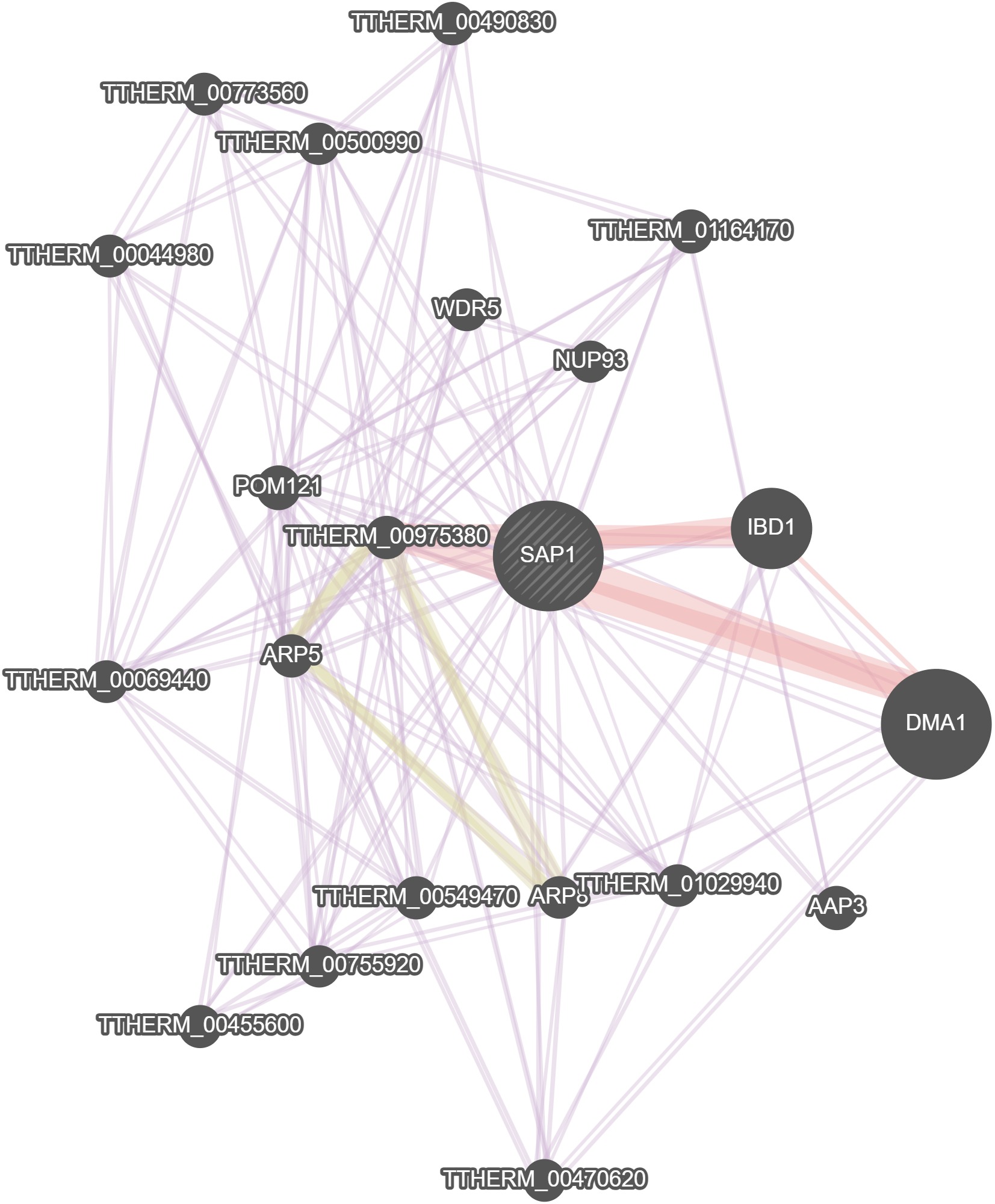
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences