Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00355870Standard Name
SEC1 (SECretory )Aliases
PreTt27573 | 37.m00290 | 3836.m02379Description
SEC1 Sec1 family protein; Sm-like protein involved in docking and fusion of exocytic vesicles; Sec1-like proteinGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- alpha6-beta1 integrin-CD151 complex (IEA) | GO:0071059
- alpha6-beta4 integrin-Shc-Grb2 complex (IEA) | GO:0070333
- biofilm matrix (IEA) | GO:0062039
- cell cortex of growing cell tip (IEA) | GO:1902716
- cellulosome (IEA) | GO:0043263
- chloroplast ribosome (IEA) | GO:0043253
- cytochrome complex (IEA) | GO:0070069
- dense core granule membrane (IEA) | GO:0032127
- EGFR-Grb2-Sos complex (IEA) | GO:0070620
- inner plaque of mitotic spindle pole body (IEA) | GO:0061497
- laminin-8 complex (IEA) | GO:0043257
- lateral plasma membrane (IEA) | GO:0016328
- low-density lipoprotein receptor complex (IEA) | GO:0062136
- MAML2-RBP-Jkappa-ICN1 complex (IEA) | GO:0071175
- obsolete chromosome segregation-directing complex (IEA) | GO:0043913
- obsolete ectoplasm (IEA) | GO:0043265
- obsolete plasma membrane part of cell junction (IEA) | GO:0061466
- outer plaque of mitotic spindle pole body (IEA) | GO:0061499
- syntaxin-6-syntaxin-16-Vti1a complex (IEA) | GO:0070067
- THO complex (IEA) | GO:0000347
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-octaprenyl-6-methoxyphenol hydroxylase activity (IEA) | GO:0008681
- 3-hydroxybutyrate dehydrogenase activity (IEA) | GO:0003858
- Ac-Asp-Glu binding (IEA) | GO:1904492
- amino acid transmembrane transporter activity (IEA) | GO:0015171
- arsenate reductase (glutaredoxin) activity (IEA) | GO:0008794
- calcium:monoatomic cation antiporter activity (IEA) | GO:0015368
- catechol O-methyltransferase activity (IEA) | GO:0016206
- Cys-tRNA(Pro) hydrolase activity (IEA) | GO:0043907
- D-loop DNA binding (IEA) | GO:0062037
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- ferredoxin-NAD(P) reductase activity (IEA) | GO:0008937
- FMN reductase (NADH) activity (IEA) | GO:0052874
- folate:monoatomic anion antiporter activity (IEA) | GO:0008518
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- growth hormone-releasing hormone receptor activity (IEA) | GO:0016520
- inositol hexakisphosphate 2-phosphatase activity (IEA) | GO:0052826
- inositol monophosphate phosphatase activity (IEA) | GO:0052834
- insulin-like growth factor II binding (IEA) | GO:0031995
- kappa-type opioid receptor binding (IEA) | GO:0031851
- Kdo2-lipid IVA acyltransferase activity (IEA) | GO:0008913
- L-ascorbate:sodium symporter activity (IEA) | GO:0008520
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- L-serine transporter activity (IEA) | GO:1905361
- lactaldehyde reductase activity (IEA) | GO:0008912
- leucyl-tRNA--protein transferase activity (IEA) | GO:0008914
- lipoate-protein ligase A activity (IEA) | GO:0008916
- lipopolysaccharide glucosyltransferase I activity (IEA) | GO:0008919
- lipopolysaccharide heptosyltransferase activity (IEA) | GO:0008920
- lipopolysaccharide-1,6-galactosyltransferase activity (IEA) | GO:0008921
- lysophosphatidic acid binding (IEA) | GO:0035727
- mannitol-1-phosphate 5-dehydrogenase activity (IEA) | GO:0008926
- mercury ion transmembrane transporter activity (IEA) | GO:0015097
- microtubule severing ATPase activity (IEA) | GO:0008568
- N-acetyl-beta-D-glucosaminide beta-(1,3)-galactosyltransferase activity (IEA) | GO:0008499
- NAD+ synthase activity (IEA) | GO:0008795
- obsolete aminoacetone:oxygen oxidoreductase(deaminating) activity (IEA) | GO:0052594
- obsolete aspartic-type signal peptidase activity (IEA) | GO:0009049
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete D-lysine oxidase activity (IEA) | GO:0043912
- obsolete kinesin motor activity (IEA) | GO:0016326
- obsolete myosin ATPase activity (IEA) | GO:0008570
- obsolete peroxisome-assembly ATPase activity (IEA) | GO:0008573
- obsolete phosphatidylinositol 3-kinase activity, class I (IEA) | GO:0016304
- obsolete phosphatidylinositol 3-kinase, class I, catalyst activity (IEA) | GO:0015072
- obsolete proprotein convertase 2 activator activity (IEA) | GO:0016484
- obsolete RNA polymerase I transcription regulator recruiting activity (IEA) | GO:0001186
- obsolete RNA polymerase III transcription factor activity (IEA) | GO:0003709
- obsolete sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity (IEA) | GO:0052591
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity (IEA) | GO:0016314
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxylase activity (IEA) | GO:0008964
- phosphofructokinase activity (IEA) | GO:0008443
- pituitary adenylate cyclase activating polypeptide activity (IEA) | GO:0016521
- protein arginine kinase activity (IEA) | GO:1990424
- pyrimidine nucleotide-sugar transmembrane transporter activity (IEA) | GO:0015165
- replication fork barrier binding (IEA) | GO:0031634
- ribose 1,5-bisphosphate isomerase activity (IEA) | GO:0043917
- secretin receptor activity (IEA) | GO:0015055
- Ser(Gly)-tRNA(Ala) hydrolase activity (IEA) | GO:0043908
- sphingosine hydroxylase activity (IEA) | GO:0000170
- taste receptor binding (IEA) | GO:0031883
- threonine-type peptidase activity (IEA) | GO:0070003
- thrombospondin receptor activity (IEA) | GO:0070053
- transcription coregulator activity (IEA) | GO:0003712
- type 3 somatostatin receptor binding (IEA) | GO:0031880
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
- type 5 somatostatin receptor binding (IEA) | GO:0031882
- zinc efflux active transmembrane transporter activity (IEA) | GO:0015341
Biological Process
- (R)-2-hydroxy-alpha-linolenic acid biosynthetic process (IEA) | GO:1902609
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- activated CD8-positive, alpha-beta T cell apoptotic process (IEA) | GO:1905397
- allantoate transport (IEA) | GO:0015719
- antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent (IEA) | GO:0002489
- apolipoprotein E recycling (IEA) | GO:0071828
- apoptotic mitochondrial changes (IEA) | GO:0008637
- apoptotic process involved in heart morphogenesis (IEA) | GO:0003278
- apoptotic process involved in heart valve morphogenesis (IEA) | GO:0003276
- aspartate family amino acid biosynthetic process (IEA) | GO:0009067
- atrial septum intermedium development (IEA) | GO:0003286
- atrial septum primum morphogenesis (IEA) | GO:0003289
- baculum development (IEA) | GO:1990375
- baroreceptor detection of decreased arterial stretch (IEA) | GO:0003024
- bilirubin conjugation (IEA) | GO:0006789
- branch elongation involved in mammary gland duct branching (IEA) | GO:0060751
- branching involved in mammary gland duct morphogenesis (IEA) | GO:0060444
- branching involved in prostate gland morphogenesis (IEA) | GO:0060442
- budding cell apical bud growth (IEA) | GO:0007118
- budding cell bud growth (IEA) | GO:0007117
- budding cell isotropic bud growth (IEA) | GO:0007119
- cardiac septum development (IEA) | GO:0003279
- carotenoid metabolic process (IEA) | GO:0016116
- CD8-positive, alpha-beta cytotoxic T cell differentiation (IEA) | GO:0002308
- cellular response to dopamine (IEA) | GO:1903351
- cellular response to flavonoid (IEA) | GO:1905396
- cellular response to organic cyclic compound (IEA) | GO:0071407
- chronic inflammatory response to antigenic stimulus (IEA) | GO:0002439
- collagen biosynthetic process (IEA) | GO:0032964
- collagen metabolic process (IEA) | GO:0032963
- commissural neuron axon guidance (IEA) | GO:0071679
- common myeloid progenitor cell proliferation (IEA) | GO:0035726
- cytotoxic T cell pyroptotic cell death (IEA) | GO:1902483
- detection of stimulus involved in Dma1-dependent checkpoint (IEA) | GO:1902419
- detection of stimulus involved in sensory perception of pain (IEA) | GO:0062149
- dichotomous subdivision of terminal units involved in lung branching (IEA) | GO:0060448
- DNA replication preinitiation complex assembly (IEA) | GO:0071163
- embryonic brain development (IEA) | GO:1990403
- endothelium development (IEA) | GO:0003158
- epithelium regeneration (IEA) | GO:1990399
- ergosteryl 3-beta-D-glucoside catabolic process (IEA) | GO:1904462
- establishment or maintenance of monopolar cell polarity (IEA) | GO:0061339
- establishment or maintenance of monopolar cell polarity regulating cell shape (IEA) | GO:0061340
- extraembryonic membrane development (IEA) | GO:1903867
- fatty acid catabolic process (IEA) | GO:0009062
- fatty-acyl-CoA transport (IEA) | GO:0015916
- fibroblast growth factor receptor signaling pathway (IEA) | GO:0008543
- flocculation via extracellular polymer (IEA) | GO:0032128
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- gamma-tubulin complex assembly (IEA) | GO:1902481
- glycolytic process from glycerol (IEA) | GO:0061613
- glycolytic process through fructose-6-phosphate (IEA) | GO:0061615
- hair cycle process (IEA) | GO:0022405
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- interleukin-12-mediated signaling pathway (IEA) | GO:0035722
- interleukin-15-mediated signaling pathway (IEA) | GO:0035723
- interleukin-16 secretion (IEA) | GO:0072614
- interleukin-18 secretion (IEA) | GO:0072616
- intraciliary anterograde transport (IEA) | GO:0035720
- intraciliary retrograde transport (IEA) | GO:0035721
- L-alanine catabolic process (IEA) | GO:0042853
- laminaritriose transport (IEA) | GO:2001097
- leucine import (IEA) | GO:0060356
- lipid homeostasis (IEA) | GO:0055088
- lipoxin biosynthetic process (IEA) | GO:2001301
- lipoxin metabolic process (IEA) | GO:2001300
- lung growth (IEA) | GO:0060437
- macromolecule biosynthetic process (IEA) | GO:0009059
- maintenance of blood vessel diameter homeostasis by renin-angiotensin (IEA) | GO:0002034
- maltoheptaose catabolic process (IEA) | GO:2001123
- maltotetraose transport (IEA) | GO:2001099
- mammary gland morphogenesis (IEA) | GO:0060443
- mitochondrial membrane organization (IEA) | GO:0007006
- mitotic cytokinetic cell separation (IEA) | GO:1902409
- mitotic nuclear division (IEA) | GO:0007067
- modulation by organism of innate immune response in other organism involved in symbiotic interaction (IEA) | GO:0052306
- mRNA splicing, via endonucleolytic cleavage and ligation (IEA) | GO:0070054
- muscle contraction (IEA) | GO:0006936
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity (IEA) | GO:0062003
- negative regulation of antigen processing and presentation of lipid antigen via MHC class Ib (IEA) | GO:0002599
- negative regulation of antimicrobial peptide production (IEA) | GO:0002785
- negative regulation of antisense RNA transcription (IEA) | GO:0060195
- negative regulation of axon guidance (IEA) | GO:1902668
- negative regulation of cell cycle phase transition (IEA) | GO:1901988
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of central B cell deletion (IEA) | GO:0002899
- negative regulation of chemokine-mediated signaling pathway (IEA) | GO:0070100
- negative regulation of DNA amplification (IEA) | GO:1904524
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of endosomal vesicle fusion (IEA) | GO:1905362
- negative regulation of epithelial cell proliferation involved in lung morphogenesis (IEA) | GO:2000795
- negative regulation of exonuclease activity (IEA) | GO:1905778
- negative regulation of glycine secretion, neurotransmission (IEA) | GO:1904625
- negative regulation of interferon-gamma secretion (IEA) | GO:1902714
- negative regulation of interleukin-10 secretion (IEA) | GO:2001180
- negative regulation of interleukin-4-mediated signaling pathway (IEA) | GO:1902215
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of neuronal signal transduction (IEA) | GO:1902848
- negative regulation of neutrophil migration (IEA) | GO:1902623
- negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway (IEA) | GO:1902176
- negative regulation of peripheral B cell deletion (IEA) | GO:0002909
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phospholipid translocation (IEA) | GO:0061093
- negative regulation of phytoalexin metabolic process (IEA) | GO:0052321
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of protein maturation (IEA) | GO:1903318
- negative regulation of protein processing (IEA) | GO:0010955
- negative regulation of receptor localization to synapse (IEA) | GO:1902684
- negative regulation of response to drug (IEA) | GO:2001024
- negative regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process (IEA) | GO:0062026
- negative regulation of tetrapyrrole biosynthetic process (IEA) | GO:1901464
- negative regulation of transcription regulatory region DNA binding (IEA) | GO:2000678
- negative regulation of vasculogenesis (IEA) | GO:2001213
- neutral lipid metabolic process (IEA) | GO:0006638
- nitrate transmembrane transport (IEA) | GO:0015706
- obsolete cellular response to Dizocilpine (IEA) | GO:1905107
- obsolete cellular response to ouabain (IEA) | GO:1905105
- obsolete dynamin family protein polymerization involved in mitochondrial fission (IEA) | GO:0003374
- obsolete histone succinylation (IEA) | GO:0106077
- obsolete Immune memory response (IEA) | GO:1990982
- obsolete induction by virus of host cell-cell fusion (IEA) | GO:0006948
- obsolete intracellular sequestering of copper ion (IEA) | GO:1902720
- obsolete MAPK cascade involved in axon regeneration (IEA) | GO:1903616
- obsolete modification by host of symbiont cytoskeleton (IEA) | GO:0052335
- obsolete movement on or near symbiont (IEA) | GO:0052194
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete negative chemotaxis in other organism involved in symbiotic interaction (IEA) | GO:0052239
- obsolete negative regulation of cGMP catabolic process (IEA) | GO:0030830
- obsolete negative regulation of histone H3-K4 trimethylation (IEA) | GO:1905436
- obsolete negative regulation of intracellular calcium activated chloride channel activity (IEA) | GO:1902939
- obsolete pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction (IEA) | GO:0052308
- obsolete positive chemotaxis in other organism involved in symbiotic interaction (IEA) | GO:0052240
- obsolete positive regulation by host of symbiont defense response (IEA) | GO:0052197
- obsolete positive regulation by organism of defense-related symbiont reactive oxygen species production (IEA) | GO:0052349
- obsolete positive regulation by organism of symbiont innate immunity (IEA) | GO:0052515
- obsolete positive regulation of nucleic acid-templated transcription (IEA) | GO:1903508
- obsolete regulation of 1-deoxy-D-xylulose-5-phosphate synthase activity (IEA) | GO:1902395
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete regulation of intracellular calcium activated chloride channel activity (IEA) | GO:1902938
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete reproduction (IEA) | GO:0000003
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- obsolete sequestering of copper ion (IEA) | GO:1902718
- obsolete sodium/potassium transport (IEA) | GO:0006815
- ommochrome biosynthetic process (IEA) | GO:0006727
- peptidoglycan biosynthetic process (IEA) | GO:0009252
- phagocytosis (IEA) | GO:0006909
- phagocytosis, engulfment (IEA) | GO:0006911
- phenanthrene catabolic process via trans-9(R),10(R)-dihydrodiolphenanthrene (IEA) | GO:0018956
- phenanthrene catabolic process via trans-9(S),10(S)-dihydrodiolphenanthrene (IEA) | GO:0018957
- photosynthetic NADP+ reduction (IEA) | GO:0009780
- plant-type cell wall modification (IEA) | GO:0009827
- plasma lipoprotein particle disassembly (IEA) | GO:0071829
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of actin binding (IEA) | GO:1904618
- positive regulation of antifungal innate immune response (IEA) | GO:1905036
- positive regulation of antigen processing and presentation of peptide antigen via MHC class Ib (IEA) | GO:0002597
- positive regulation of antimicrobial peptide biosynthetic process (IEA) | GO:0002807
- positive regulation of antimicrobial peptide secretion (IEA) | GO:0002796
- positive regulation of ATF6-mediated unfolded protein response (IEA) | GO:1903893
- positive regulation of cell cycle phase transition (IEA) | GO:1901989
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of forebrain neuron differentiation (IEA) | GO:2000979
- positive regulation of hindgut contraction (IEA) | GO:0060450
- positive regulation of inner ear receptor cell differentiation (IEA) | GO:2000982
- positive regulation of inositol trisphosphate biosynthetic process (IEA) | GO:0032962
- positive regulation of interleukin-10 secretion (IEA) | GO:2001181
- positive regulation of meiotic cell cycle phase transition (IEA) | GO:1901995
- positive regulation of myofibroblast differentiation (IEA) | GO:1904762
- positive regulation of ovulation (IEA) | GO:0060279
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway (IEA) | GO:1902177
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of plant epidermal cell differentiation (IEA) | GO:1903890
- positive regulation of skeletal muscle satellite cell differentiation (IEA) | GO:1902726
- positive regulation of tumor necrosis factor biosynthetic process (IEA) | GO:0042535
- positive regulation of venous endothelial cell fate commitment (IEA) | GO:2000789
- pre-replicative complex assembly involved in cell cycle DNA replication (IEA) | GO:1902299
- programmed cell death (IEA) | GO:0012501
- pronephric glomerulus morphogenesis (IEA) | GO:0035775
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- prostate induction (IEA) | GO:0060514
- protein lipoylation (IEA) | GO:0009249
- protein localization to meiotic spindle midzone (IEA) | GO:1903096
- protein localization to mitotic spindle (IEA) | GO:1902480
- protein localization to old growing cell tip (IEA) | GO:1903858
- proton-transporting V-type ATPase complex assembly (IEA) | GO:0070070
- pteridine biosynthetic process (IEA) | GO:0006728
- radial glial cell fate commitment in forebrain (IEA) | GO:0022023
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of actomyosin contractile ring contraction (IEA) | GO:0031991
- regulation of antibacterial peptide biosynthetic process (IEA) | GO:0002808
- regulation of antigen processing and presentation of lipid antigen via MHC class Ib (IEA) | GO:0002598
- regulation of antisense RNA transcription (IEA) | GO:0060194
- regulation of biosynthetic process of antibacterial peptides active against Gram-positive bacteria (IEA) | GO:0002816
- regulation of capsule polysaccharide biosynthetic process (IEA) | GO:0062084
- regulation of cardioblast proliferation (IEA) | GO:0003264
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of dendrite extension (IEA) | GO:1903859
- regulation of double fertilization forming a zygote and endosperm (IEA) | GO:0080155
- regulation of eIF2 alpha phosphorylation by amino acid starvation (IEA) | GO:0060733
- regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation (IEA) | GO:0060734
- regulation of exonuclease activity (IEA) | GO:1905777
- regulation of frizzled signaling pathway (IEA) | GO:0008590
- regulation of fucose catabolic process (IEA) | GO:0043468
- regulation of hemicellulose catabolic process (IEA) | GO:2000988
- regulation of indoleacetic acid biosynthetic process via tryptophan (IEA) | GO:1901996
- regulation of interferon-gamma secretion (IEA) | GO:1902713
- regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070107
- regulation of membrane lipid metabolic process (IEA) | GO:1905038
- regulation of microglial cell migration (IEA) | GO:1904139
- regulation of mitophagy (IEA) | GO:1901524
- regulation of NAD metabolic process (IEA) | GO:1902688
- regulation of PERK-mediated unfolded protein response (IEA) | GO:1903897
- regulation of phosphatidate phosphatase activity (IEA) | GO:1903730
- regulation of phosphatidylcholine biosynthetic process (IEA) | GO:2001245
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of polysaccharide metabolic process (IEA) | GO:0032881
- regulation of protein localization to cell cortex (IEA) | GO:1904776
- regulation of receptor localization to synapse (IEA) | GO:1902683
- regulation of response to cell cycle checkpoint signaling (IEA) | GO:1902145
- regulation of rhamnose catabolic process (IEA) | GO:0043463
- regulation of satellite cell activation involved in skeletal muscle regeneration (IEA) | GO:0014717
- regulation of secondary heart field cardioblast proliferation (IEA) | GO:0003266
- regulation of systemic arterial blood pressure by baroreceptor feedback (IEA) | GO:0003025
- regulation of systemic arterial blood pressure by carotid body chemoreceptor signaling (IEA) | GO:0003027
- regulation of transcriptional start site selection at RNA polymerase II promoter (IEA) | GO:0001178
- regulation of ubiquitin homeostasis (IEA) | GO:0010993
- regulation of xylan catabolic process (IEA) | GO:2001000
- response to curcumin (IEA) | GO:1904643
- response to oxygen radical (IEA) | GO:0000305
- response to reactive oxygen species (IEA) | GO:0000302
- response to simvastatin (IEA) | GO:1903491
- septal cell proliferation (IEA) | GO:0022022
- serine family amino acid biosynthetic process (IEA) | GO:0009070
- serotonin secretion involved in inflammatory response (IEA) | GO:0002442
- smooth muscle contraction (IEA) | GO:0006939
- sterol metabolic process (IEA) | GO:0016125
- striated muscle myosin thick filament assembly (IEA) | GO:0071688
- terpenoid catabolic process (IEA) | GO:0016115
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- tetrasaccharide transport (IEA) | GO:2001098
- transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript (IEA) | GO:0001180
- tRNA import into nucleus (IEA) | GO:0035719
- vacuolar acidification (IEA) | GO:0007035
- vacuolar proton-transporting V-type ATPase complex assembly (IEA) | GO:0070072
- ventricular septum intermedium development (IEA) | GO:0003282
- xanthophyll biosynthetic process (IEA) | GO:0016123
- xanthophyll catabolic process (IEA) | GO:0016124
 Domains
Domains
- ( PF00995 ) Sec1 family
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
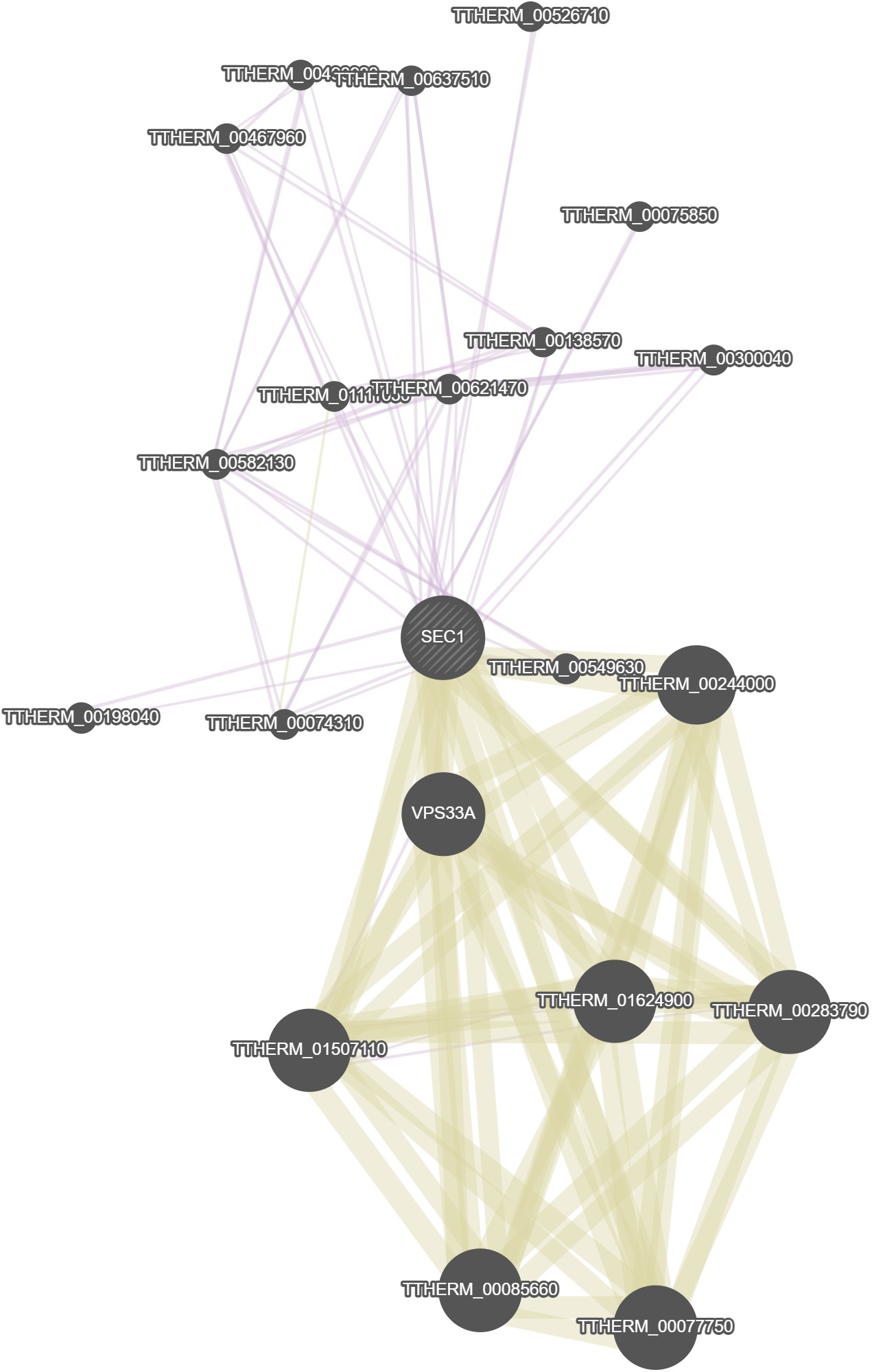
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00355870(coding)
ATGCAAATACTACAAAAACTAAATGAGAATATCAGTTAGTATTCCCTGAAAGAAAATGTT
TTCTACAGGTTTTAAGAAATGTTTGATGATGTAAGAAGCAGATACCCAAATATAGGAACA
TTTATTCTTGTTGTAGATTAGCAAAGTTTAAAACTGCTTTCTTCAGCACTTAAAATGGCA
GATTTAATAAAAATGGGTGTTTCTTCAATTGAAAAGCTGGAGTTAAAAAGAAAAGAACAT
CCAAATACGCATGCCATTTACTTACTTTCACCTAGTAAAGAATCGATAAGTCTACTTATA
TCTGACTTTGAGGGACAGGTTTTGTAACTTAAAAACACTGATGAAAAGAAAAAGAAGAAA
GCATAGAAGCAAAAGGAAAAGGAAAAGAAGAAGAAGAAGAAAAAAGGTAAACAGGACGAT
GATGATGAAGACGAATAAGAAGATGATGAAGATGATGATGAAGTTATTGAAGAAAATGAA
GAAAGCAAAAAGAAGAGGAAACAAGATGAATTAGAACCTAAAAAACCAGAGAAAAAAAAG
GATTTGCAACCACCAGAAATATAATATGCTAGAGTTCATATTTTCTTTATGAATAAGCTA
CCAAAATACTTGCTCGAAGAAATGGGAAAGTCTGAAAAGTTAGTCAAAAGAATTAGAACC
ATGAAAGAATTTAATCATTCTTTCTTTTTCTTCGAATAAAACGGATTTCATCTACATTTA
GAGTAATCTTTACCAATTCTTTTTTCTAAGAAGGAAAGCTAAGAGGCAAGAATAATGTTT
GAATGCATTTCTGATCAATTATCTACAGTTCTACCTGCTCTATTGAGGTTTGATAAGATA
AATATATTATACAATAACTAAAATCAAAGCAGTCCAAGCTATTTGTTTGCCAACTATTTT
GAAAAAAAATTGCAGGAAATGATTTTGAAATTGAAGGAAAGTGAGTCTCCATACATCGAT
CCAACATCAGGAGAGATTTACTTGATTATAGTTGACAGAGGTATAGATCCCGTCACACCT
CTTTTGCATGATTATTCTTACTAGAGCATGATTTATGATTTATTAGAAGTCGATACAGAA
AAAAATATAGTTGAGTATGAAAAAATAGAACTCAAAAAAGAAAACAAAGCAAGTGAATCT
ACAAAAGAAGTAAAAAAATATAAATAATAGCTAAGTGATCATAAGGATGAAGTTTTTGCA
AAGTTTAGATATCAAAACATCACTGAAGCTTTGAAAGGAATCAGAGATGATTTTGAAAAA
CACACAGAAAATATGTAGAGAGCAAAGTCTTCTCAAAATTAAGTTAGGAGTATTGAAGAT
GTTAACAAAATTATTGGAGAGTTTCCAGAATACAATGAACTTTTATAAAAATATGCAGTG
CATTTTGACTTGATTTTAAAATGTTTCAATACCTTTTAAAAATATAATTTGCAATTAAAC
TGTGAGCTTGAGTAATCTATGGTAACTGGAATTGATGATCTTGCAAAACCTATTAAAAGT
GATACTATTATTCAAAGTATATAAGATATATTAAATAGAAAAGATATAAAAAGCACAGAT
AGACTTAGGATAGCAATGATTGCTTTTATAACATGTGATCTCCAATAAGACTAAAGACAA
AAAATTTTAAATCTATTTGAATTAAGTGAATAAAAATAATTAAAAAACTTAGCATGGCTT
GGTATCTAATTTAGTGATGATAAAAATTCTACTTTTAGTAGAGTTAATTAACAAATTATA
GATTTATCTAAGGAGTAGTATAATGCTTATTTGAATAAATAAAAAAATTGCTAAGAATGT
AAGAAAATACGTGCAGATGCAAAAAAATCAAAGAAAGGAGGAAATTCTGAAGAGGACTAA
AAGCGAGATTGTGATGAATGTGGTTAGTTGTATTTCGTCTTTAGCGTTATGAGGCACACT
CCTCTATCAGCTATAGCTATACATTCTGCAATCAAATAAATAATTAAATCATACGATGTT
TCAAAGGATGCATTAAATAGTTTAGGGTTTGGGATAAAATAAGTTGGATAAGTTAATAGA
CCTATATCTCCACTTAAAGAATAGCCACTATTGAATGAAGAAAGTGAGATGTAATCTGAA
AATAAATTGCAATCAACAAATTCAAGTAAACAAGTAAAATCATCTAAAAAGAAAGATAAA
GACAAAAAAAAAGACAAAGAAGAATCTAAATCATAAAAAAAAGAAAAAGGTGATTTGCAT
TTACCCAATGAATATGAAGCATTTTTGTAATAATCAGAGAGTGAAGACTCAGATGCTCTC
AAAAAAAGTAAAAAAATAATTGCTTTCTTTATTGGTGGAATCAGTTATGCAGAAATTAGA
GCTATCAGAAAATTCTAGGATGAATATTAGAGTAATCCAGTAATAATAGGAGCAACTCAC
TTGCTTACCCATCGCAACTACATAGAAGAGATAGACCAAATGTCAACTTTTGCTTGA>TTHERM_00355870(gene)
TTTTAATATTTCAAGCTTAGCAAATAATAAATAAAAAAATCAGTATCAATCAACCAGCCA
TTTTATAGCAATTAGTCAATTAGTAAAAATAAAAGAAAAGAGTTCAAATATTATTTATAG
CAACTTAGAACATTTTTAAAAGAACTTATAATGCAAATACTACAAAAACTAAATGAGAAT
ATCAGTTAGTATTCCCTGAAAGAAAATGTTTTCTACAGGTTTTAAGAAATGTTTGATGAT
GTAAGAAGCAGATACCCAAATATAGGAACATTTATTCTTGTTGTAGATTAGCAAAGTTTA
AAACTGCTTTCTTCAGCACTTAAAATGGCAGATTTAATAAAAATGGGTGTTTCTTCAATT
GAAAAGCTGGAGTTAAAAAGAAAAGAACATCCAAATACGCATGCCATTTACTTACTTTCA
CCTAGTAAAGAATCGATAAGTCTACTTATATCTGACTTTGAGGGACAGGTTTTGTAACTT
AAAAACACTGATGAAAAGAAAAAGAAGAAAGCATAGAAGCAAAAGGAAAAGGAAAAGAAG
AAGAAGAAGAAAAAAGGTAAACAGGACGATGATGATGAAGACGAATAAGAAGATGATGAA
GATGATGATGAAGTTATTGAAGAAAATGAAGAAAGCAAAAAGAAGAGGAAACAAGATGAA
TTAGAACCTAAAAAACCAGAGAAAAAAAAGGATTTGCAACCACCAGAAATATAATATGCT
AGAGTTCATATTTTCTTTATGAATAAGCTACCAAAATACTTGCTCGAAGAAATGGGAAAG
TCTGAAAAGTTAGTCAAAAGAATTAGAACCATGAAAGAATTTAATCATTCTTTCTTTTTC
TTCGAATAAAACGGATTTCATCTACATTTAGAGTAATCTTTACCAATTCTTTTTTCTAAG
AAGGAAAGCTAAGAGGCAAGAATAATGTTTGAATGCATTTCTGATCAATTATCTACAGTT
CTACCTGCTCTATTGAGGTTTGATAAGATAAATATATTATACAATAACTAAAATCAAAGC
AGTCCAAGCTATTTGTTTGCCAACTATTTTGAAAAAAAATTGCAGGAAATGATTTTGAAA
TTGAAGGAAAGTGAGTCTCCATACATCGATCCAACATCAGGAGAGATTTACTTGATTATA
GTTGACAGAGGTATAGATCCCGTCACACCTCTTTTGCATGATTATTCTTACTAGAGCATG
ATTTATGATTTATTAGAAGTCGATACAGAAAAAAATATAGTTGAGTATGAAAAAATAGAA
CTCAAAAAAGAAAACAAAGCAAGTGAATCTACAAAAGAAGTAAAAAAATATAAATAATAG
CTAAGTGATCATAAGGATGAAGTTTTTGCAAAGTTTAGATATCAAAACATCACTGAAGCT
TTGAAAGGAATCAGAGATGATTTTGAAAAACACACAGAAAATATGTAGAGAGCAAAGTCT
TCTCAAAATTAAGTTAGGAGTATTGAAGATGTTAACAAAATTATTGGAGAGTTTCCAGAA
TACAATGAACTTTTATAAAAATATGCAGTGCATTTTGACTTGATTTTAAAATGTTTCAAT
ACCTTTTAAAAATATAATTTGCAATTAAACTGTGAGCTTGAGTAATCTATGGTAACTGGA
ATTGATGATCTTGCAAAACCTATTAAAAGTGATACTATTATTCAAAGTATATAAGATATA
TTAAATAGAAAAGATATAAAAAGCACAGATAGACTTAGGATAGCAATGATTGCTTTTATA
ACATGTGATCTCCAATAAGACTAAAGACAAAAAATTTTAAATCTATTTGAATTAAGTGAA
TAAAAATAATTAAAAAACTTAGCATGGCTTGGTATCTAATTTAGTGATGATAAAAATTCT
ACTTTTAGTAGAGTTAATTAACAAATTATAGATTTATCTAAGGAGTAGTATAATGCTTAT
TTGAATAAATAAAAAAATTGCTAAGAATGTAAGAAAATACGTGCAGATGCAAAAAAATCA
AAGAAAGGAGGAAATTCTGAAGAGGACTAAAAGCGAGATTGTGATGAATGTGGTTAGTTG
TATTTCGTCTTTAGCGTTATGAGGCACACTCCTCTATCAGCTATAGCTATACATTCTGCA
ATCAAATAAATAATTAAATCATACGATGTTTCAAAGGATGCATTAAATAGTTTAGGGTTT
GGGATAAAATAAGTTGGATAAGTTAATAGACCTATATCTCCACTTAAAGAATAGCCACTA
TTGAATGAAGAAAGTGAGATGTAATCTGAAAATAAATTGCAATCAACAAATTCAAGTAAA
CAAGTAAAATCATCTAAAAAGAAAGATAAAGACAAAAAAAAAGACAAAGAAGAATCTAAA
TCATAAAAAAAAGAAAAAGGTGATTTGCATTTACCCAATGAATATGAAGCATTTTTGTAA
TAATCAGAGAGTGAAGACTCAGATGCTCTCAAAAAAAGTAAAAAAATAATTGCTTTCTTT
ATTGGTGGAATCAGTTATGCAGAAATTAGAGCTATCAGAAAATTCTAGGATGAATATTAG
AGTAATCCAGTAATAATAGGAGCAACTCACTTGCTTACCCATCGCAACTACATAGAAGAG
ATAGACCAAATGTCAACTTTTGCTTGACTGGTTTTTAATCAATAAATAAATTAGAATTTA
ATGATTTTATTATCTTGATCTCAATTTAATAGTTCAAAAATAAATAAATTCTAAAACTAA
ATATATCAAAAAATAAATAAATATTATCAGAAAATTATTTCATTTTATTAAAAAAAAAGA
ATTTAAGTTTTGGCTTTGACCTTTGTAAATTACATAATATATGATGAAAAAAAGTAATTT
AAAAATTAAATGGAAAT>TTHERM_00355870(protein)
MQILQKLNENISQYSLKENVFYRFQEMFDDVRSRYPNIGTFILVVDQQSLKLLSSALKMA
DLIKMGVSSIEKLELKRKEHPNTHAIYLLSPSKESISLLISDFEGQVLQLKNTDEKKKKK
AQKQKEKEKKKKKKKGKQDDDDEDEQEDDEDDDEVIEENEESKKKRKQDELEPKKPEKKK
DLQPPEIQYARVHIFFMNKLPKYLLEEMGKSEKLVKRIRTMKEFNHSFFFFEQNGFHLHL
EQSLPILFSKKESQEARIMFECISDQLSTVLPALLRFDKINILYNNQNQSSPSYLFANYF
EKKLQEMILKLKESESPYIDPTSGEIYLIIVDRGIDPVTPLLHDYSYQSMIYDLLEVDTE
KNIVEYEKIELKKENKASESTKEVKKYKQQLSDHKDEVFAKFRYQNITEALKGIRDDFEK
HTENMQRAKSSQNQVRSIEDVNKIIGEFPEYNELLQKYAVHFDLILKCFNTFQKYNLQLN
CELEQSMVTGIDDLAKPIKSDTIIQSIQDILNRKDIKSTDRLRIAMIAFITCDLQQDQRQ
KILNLFELSEQKQLKNLAWLGIQFSDDKNSTFSRVNQQIIDLSKEQYNAYLNKQKNCQEC
KKIRADAKKSKKGGNSEEDQKRDCDECGQLYFVFSVMRHTPLSAIAIHSAIKQIIKSYDV
SKDALNSLGFGIKQVGQVNRPISPLKEQPLLNEESEMQSENKLQSTNSSKQVKSSKKKDK
DKKKDKEESKSQKKEKGDLHLPNEYEAFLQQSESEDSDALKKSKKIIAFFIGGISYAEIR
AIRKFQDEYQSNPVIIGATHLLTHRNYIEEIDQMSTFA