 Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00388500
Standard Name
SWC2
(SWR complex component)
Aliases
PreTt24845 | 41.m00252
Description
SWC2 YL1 nuclear protein; Member of the SWR complex; Co-purifies with Hv1 (Kanwal Ashraf et al., 2019); hypothetical protein
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
No Data fetched for Gene Ontology Annotations
 Domains
Domains
No Data fetched for Domains
 Gene Expression Profile
Gene Expression Profile
 GeneMania
GeneMania
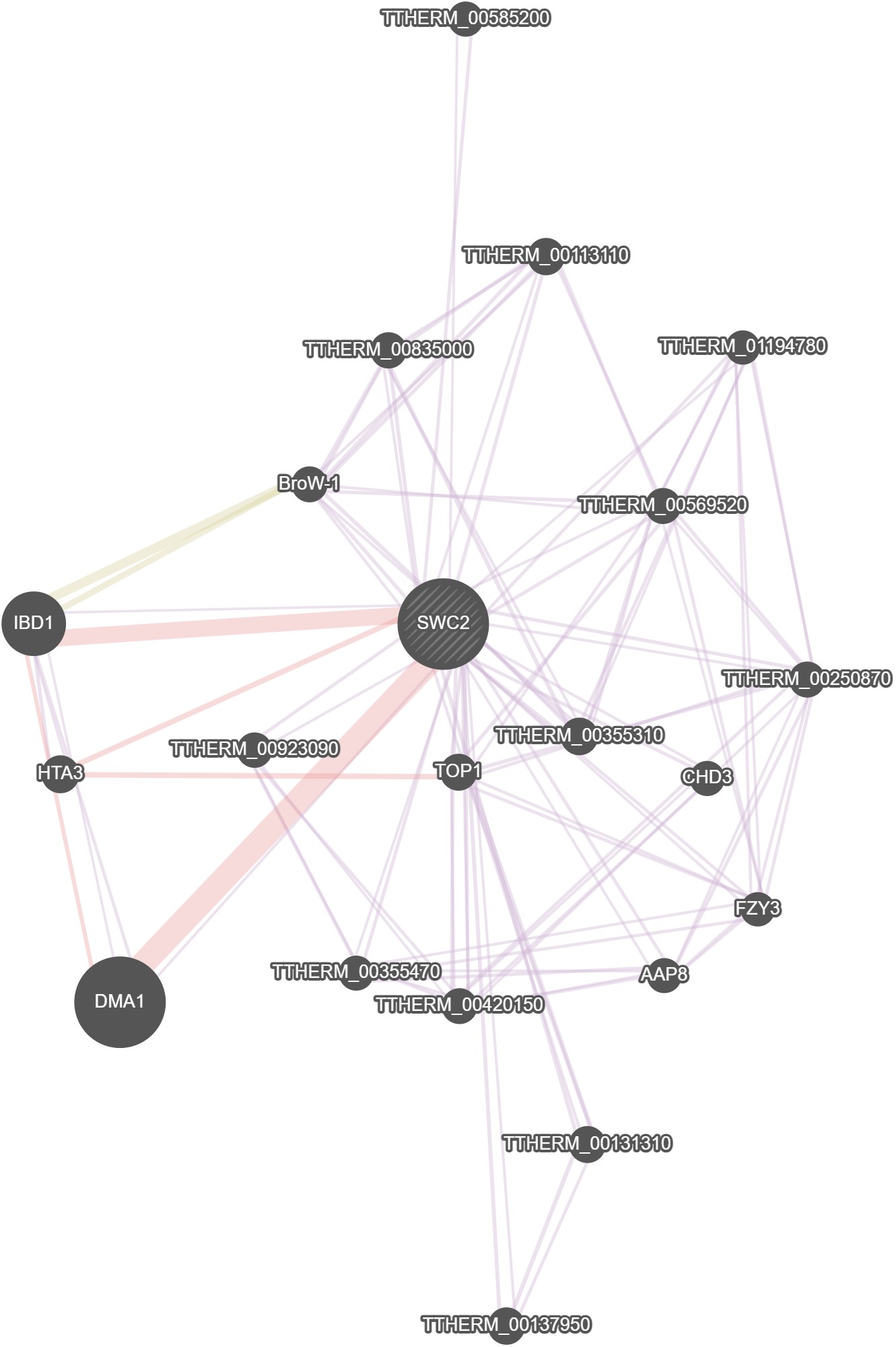
No results were calculated for this gene in GeneMania.
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:30796450: Ashraf K, Nabeel-Shah S, Garg J, Saettone A, Derynck J, Gingras AC, Lambert JP, Pearlman RE, Fillingham J (2019) Proteomic analysis of histones H2A/H2B and variant Hv1 in Tetrahymena thermophila reveals an ancient network of chaperones. Molecular biology and evolution ( ):
- Ref:29523178: Saettone A, Garg J, Lambert JP, Nabeel-Shah S, Ponce M, Burtch A, Thuppu Mudalige C, Gingras AC, Pearlman RE, Fillingham J (2018) The bromodomain-containing protein Ibd1 links multiple chromatin-related protein complexes to highly expressed genes in Tetrahymena thermophila. Epigenetics & chromatin 11(1):10
 Sequences
Sequences
>TTHERM_00388500(coding)
ATGGGAAAGAAAGAAAAGGTTTTCTTTGAGGAGGAAGATGATAACAAGAATGAGGAGATA
TATAATAGCGATTAAGATGATTAAGATTAAACTCCTTAAAACAAATCACATTCAAACAAT
TCTAATTAAGTAGGCGCATAGTCTAAAAAAGGAGCTGTTTAAGATAAATCAGCTAAAAAT
TCTAAAGTCACAGCATCAAAAGGAAATCAAAAGAAGAAATCATAAGAAGTAGATTCAAAA
GCTCCAAAGAAAGGGAAAGCTATTTCAAAAGGCAAAACTAATAAAAAAGGAAAAGACAAA
GGTGGTGCCGCTTCTAAAAAAGGAGGTAAAAAGAAAGTACAAAAAGGTAAAAAAGGAGAA
AAGGCTCCAGTGAAAGGTTCAGCTACTAAAAAAAAAGGAGGTGCTTAAGATAAGAAAAAA
GCAGATAATAGCGCTCTTAAAAAGACAAAAAAAATTTCATAGGGTAAAGCAGAAATCATA
GAAGATGAAAAAGAAGAAGTATTTAATGATGAAAGTAATGCAACTCCTCCTTAATATAGC
TAAACAAATGCTACTTCTTATTAAAGCGATAATATCTCAAAGCTAAAAAAAAAACCAGCT
GCAAAAGCTAGCAATAAAAAAACACAATAAACTCTAGCTGCATCTAAATTAAAAACGCAA
AAAGAGGAATAAGGAGAAAAATCTTCTAGTTTGAAAGCATTTAGCAGCATAATTAAAGAC
AAAAAGAATGGAAAAAACGTTCCTTTCCAGCCTACAGAAAATAAAGAGAGTGAAGAAATA
GAAATAGAACCTGAGACAGATGAATAAGAATCAGATCCTAATTAGCTTAAAAAGAAGCTA
GGTAAATCAGATTTAAATACTGAGCATCAAAATGGTGGAAATTAAGTAAAAGATGATAAA
AAAGGTTAATCTGAATCATCTTTAAAAAATAGAAGCAAGGTCTTAGTTGATAGTAAAAAA
CCTAATTCGAAGCTTTATAAAAATGATAATGGTCATGAATAAAGTACTACTTCTAAAGGG
TCTAAGGTATCTGCTATTAAAAACATAAGAAAGGATAGTTTAGATATGGAATTAGACAAT
ACTTTAGAAAACGAAGAGGATGAAGAAGATATTCTTTCAGGTTCTGAAGATTCTGAAAAT
AGCTAAAACAGTGATAATATAAAAACAAACAAAAAATAATTGGACAAGAAGAAAAGCGAT
AGTTTATCATCAAAACTTAAAAATAAAGCTGATGAAAGTGAAGATGATGGAGAAGGAGAA
GACAATGAAGAAGACGATGAAGTTGATGATGACGAAGAAGACGAAGAGAATGGTGATGAG
GATGATGATGATGACGAAAATGAAGGTGATGATGAAGAAGATAACGATGAAGACGAAAGT
AGTAGTGAAGAAGTAGTTTAGGCTGTAAGAACAATGACATTTCGTAGTAGAAGAGGTGAA
GGCATCAAAAAGCTTATGGAGTTATAAAAAAGAAATGAAGGATTCGGAGATGATTTAGAT
GACTTTTGGAAAGAAAATAAATATTTTGGTAAAATAACTGAAGTTACAAAGGCTAATGAT
GAAGAAGAAGACGAAGATTTTAATGAAGAAGAGTTTAAAGACGTAAAGGATGTATTCGAT
AGTGATTTTTAAGACACTAGTGATTTAGAAGATGATGGCGATGATGATTCAAATATGTCA
GATGAAGATTCAGACAGCGAGAAAAAGAAAAAACGTAATAAAGATAAGCCTAGCTATTTA
ATGAAAAAAGATGATGTTATTAGAAAAGAATAAGAAAAAATAGCAAAGCAGAGACAAAAA
TAAAAAGCTGAGCGCTTGAAATAGCTCGATACCTAAGAAGAAAATGAAATTTAAATATCT
TAAAAGATATAAAAGGAGAAAAGAGAGGCATAGATCACAATTTAGCTTGAAAAACGTACC
TTAAGATAAAGCACAAATGAAAAATCTCTTTTATCTCAAAGTTAGCTTTCTCCTCGTTTA
AAAAAGAAATTGCGCTTAAACGAAGATGGATCTGTAGACATTAATGGGTAATAAGGAGAG
TAAGGTGGATAAGCACAGGGTGCTGGAGGCAAAAAGAAACGTTATATCCATGATTTCTTG
ACCTAAGAGTAACTATTAAAAGAAGCTGTTTTTACAGAATTTGTGAATAGAAAATAATTA
GAAGACTTACTTCGTTTGGAAGAAGAGAAAAAATAATTCTCTTTAAGTAAAAAATTTATA
GACGAAGGACCTAAGATAATATATAAAAACTCTGCTAAAACAGATGAAAATTTAATCATA
TATCCAACATAAGAAGTATATGATAGCACAATAGGTAACTATATGTAATCTTTCAATCAA
TAAAAATAAATAAAAGAAAAAAAATTCCGCTATCACGATCCTAAAACTAATAAATCTTAT
AATACTCCAGAAGAATTTTATAAATATCGTACAATTTTGTATAAGTCAGAGCAAAATCGT
TACAAGTATCTTATTAAAGTTTACAAAGATCTCTAAGAAAAGAAGGAAGAGCTGCTAGAA
GAATTTGAGCCAAAACTTAAAGACAAATATAATAATAATTGTAATAATTCTTCAGATTCA
GATGACTCTTTATCATCATCTTCATAATCCTCATCTAATTCAGCAACTTCTTCTGCAGGT
AAAAAGAAGAAGAGAAAATCTTAATAAAAGAAAAAGAAACAAACTAAGAAAGCAAAAAGT
TAAAAATCTCGTATTTCATAAACTGCAGCTGAAAGAGAAGATGATGAAAATGATGAAGAA
AACGATGATGATAATAATAATAATTAAGATGATAAAATATTTATTGACTAAGATTCAGAT
TGA
>TTHERM_00388500(gene)
GAAATAAATAAGTAAATAAATAAATAAAGATTAATAGACCTTCGCGGATTGAAATATTTT
TTAAATTTCATTTTTAATAAATAATAAATAATAAAAATTCTTAGTTTAAAAATTGAATAT
TTGACATATTCAATATGGGAAAGAAAGAAAAGGTTTTCTTTGAGGAGGAAGATGATAACA
AGAATGAGGAGATATATAATAGCGATTAAGATGATTAAGATTAAACTCCTTAAAACAAAT
CACATTCAAACAATTCTAATTAAGTAGGCGCATAGTCTAAAAAAGGAGCTGTTTAAGATA
AATCAGCTAAAAATTCTAAAGTCACAGCATCAAAAGGAAATCAAAAGAAGAAATCATAAG
AAGTAGATTCAAAAGCTCCAAAGAAAGGGAAAGCTATTTCAAAAGGCAAAACTAATAAAA
AAGGAAAAGACAAAGGTGGTGCCGCTTCTAAAAAAGGAGGTAAAAAGAAAGTACAAAAAG
GTAAAAAAGGAGAAAAGGCTCCAGTGAAAGGTTCAGCTACTAAAAAAAAAGGAGGTGCTT
AAGATAAGAAAAAAGCAGATAATAGCGCTCTTAAAAAGACAAAAAAAATTTCATAGGGTA
AAGCAGAAATCATAGAAGATGAAAAAGAAGAAGTATTTAATGATGAAAGTAATGCAACTC
CTCCTTAATATAGCTAAACAAATGCTACTTCTTATTAAAGCGATAATATCTCAAAGCTAA
AAAAAAAACCAGCTGCAAAAGCTAGCAATAAAAAAACACAATAAACTCTAGCTGCATCTA
AATTAAAAACGCAAAAAGAGGAATAAGGAGAAAAATCTTCTAGTTTGAAAGCATTTAGCA
GCATAATTAAAGACAAAAAGAATGGAAAAAACGTTCCTTTCCAGCCTACAGAAAATAAAG
AGAGTGAAGAAATAGAAATAGAACCTGAGACAGATGAATAAGAATCAGATCCTAATTAGC
TTAAAAAGAAGCTAGGTAAATCAGATTTAAATACTGAGCATCAAAATGGTGGAAATTAAG
TAAAAGATGATAAAAAAGGTTAATCTGAATCATCTTTAAAAAATAGAAGCAAGGTCTTAG
TTGATAGTAAAAAACCTAATTCGAAGCTTTATAAAAATGATAATGGTCATGAATAAAGTA
CTACTTCTAAAGGGTCTAAGGTATCTGCTATTAAAAACATAAGAAAGGATAGTTTAGATA
TGGAATTAGACAATACTTTAGAAAACGAAGAGGATGAAGAAGATATTCTTTCAGGTTCTG
AAGATTCTGAAAATAGCTAAAACAGTGATAATATAAAAACAAACAAAAAATAATTGGACA
AGAAGAAAAGCGATAGTTTATCATCAAAACTTAAAAATAAAGCTGATGAAAGTGAAGATG
ATGGAGAAGGAGAAGACAATGAAGAAGACGATGAAGTTGATGATGACGAAGAAGACGAAG
AGAATGGTGATGAGGATGATGATGATGACGAAAATGAAGGTGATGATGAAGAAGATAACG
ATGAAGACGAAAGTAGTAGTGAAGAAGTAGTTTAGGCTGTAAGAACAATGACATTTCGTA
GTAGAAGAGGTGAAGGCATCAAAAAGCTTATGGAGTTATAAAAAAGAAATGAAGGATTCG
GAGATGATTTAGATGACTTTTGGAAAGAAAATAAATATTTTGGTAAAATAACTGAAGTTA
CAAAGGCTAATGATGAAGAAGAAGACGAAGATTTTAATGAAGAAGAGTTTAAAGACGTAA
AGGATGTATTCGATAGTGATTTTTAAGACACTAGTGATTTAGAAGATGATGGCGATGATG
ATTCAAATATGTCAGATGAAGATTCAGACAGCGAGAAAAAGAAAAAACGTAATAAAGATA
AGCCTAGCTATTTAATGAAAAAAGATGATGTTATTAGAAAAGAATAAGAAAAAATAGCAA
AGCAGAGACAAAAATAAAAAGCTGAGCGCTTGAAATAGCTCGATACCTAAGAAGAAAATG
AAATTTAAATATCTTAAAAGATATAAAAGGAGAAAAGAGAGGCATAGATCACAATTTAGC
TTGAAAAACGTACCTTAAGATAAAGCACAAATGAAAAATCTCTTTTATCTCAAAGTTAGC
TTTCTCCTCGTTTAAAAAAGAAATTGCGCTTAAACGAAGATGGATCTGTAGACATTAATG
GGTAATAAGGAGAGTAAGGTGGATAAGCACAGGGTGCTGGAGGCAAAAAGAAACGTTATA
TCCATGATTTCTTGACCTAAGAGTAACTATTAAAAGAAGCTGTTTTTACAGAATTTGTGA
ATAGAAAATAATTAGAAGACTTACTTCGTTTGGAAGAAGAGAAAAAATAATTCTCTTTAA
GTAAAAAATTTATAGACGAAGGACCTAAGATAATATATAAAAACTCTGCTAAAACAGATG
AAAATTTAATCATATATCCAACATAAGAAGTATATGATAGCACAATAGGTAACTATATGT
AATCTTTCAATCAATAAAAATAAATAAAAGAAAAAAAATTCCGCTATCACGATCCTAAAA
CTAATAAATCTTATAATACTCCAGAAGAATTTTATAAATATCGTACAATTTTGTATAAGT
CAGAGCAAAATCGTTACAAGTATCTTATTAAAGTTTACAAAGATCTCTAAGAAAAGAAGG
AAGAGCTGCTAGAAGAATTTGAGCCAAAACTTAAAGACAAATATAATAATAATTGTAATA
ATTCTTCAGATTCAGATGACTCTTTATCATCATCTTCATAATCCTCATCTAATTCAGCAA
CTTCTTCTGCAGGTAAAAAGAAGAAGAGAAAATCTTAATAAAAGAAAAAGAAACAAACTA
AGAAAGCAAAAAGTTAAAAATCTCGTATTTCATAAACTGCAGCTGAAAGAGAAGATGATG
AAAATGATGAAGAAAACGATGATGATAATAATAATAATTAAGATGATAAAATATTTATTG
ACTAAGATTCAGATTGATGATAGATGAACTTTCAACAATCAATATACTAAAAATTTTTTA
GATAAATAATTATTTGTAAATAATATTTTAAATTAAACTGTCATTTTTAGATTTTTCAAA
AATTTTAAAAAATTAATAATTTTTTTTTCTCAATTAATTAAATAAAACATTTTTAAAATA
AAATTAGTAAATATTTAATTTTTTTTCATAAAAAAATACAATT
>TTHERM_00388500(protein)
MGKKEKVFFEEEDDNKNEEIYNSDQDDQDQTPQNKSHSNNSNQVGAQSKKGAVQDKSAKN
SKVTASKGNQKKKSQEVDSKAPKKGKAISKGKTNKKGKDKGGAASKKGGKKKVQKGKKGE
KAPVKGSATKKKGGAQDKKKADNSALKKTKKISQGKAEIIEDEKEEVFNDESNATPPQYS
QTNATSYQSDNISKLKKKPAAKASNKKTQQTLAASKLKTQKEEQGEKSSSLKAFSSIIKD
KKNGKNVPFQPTENKESEEIEIEPETDEQESDPNQLKKKLGKSDLNTEHQNGGNQVKDDK
KGQSESSLKNRSKVLVDSKKPNSKLYKNDNGHEQSTTSKGSKVSAIKNIRKDSLDMELDN
TLENEEDEEDILSGSEDSENSQNSDNIKTNKKQLDKKKSDSLSSKLKNKADESEDDGEGE
DNEEDDEVDDDEEDEENGDEDDDDDENEGDDEEDNDEDESSSEEVVQAVRTMTFRSRRGE
GIKKLMELQKRNEGFGDDLDDFWKENKYFGKITEVTKANDEEEDEDFNEEEFKDVKDVFD
SDFQDTSDLEDDGDDDSNMSDEDSDSEKKKKRNKDKPSYLMKKDDVIRKEQEKIAKQRQK
QKAERLKQLDTQEENEIQISQKIQKEKREAQITIQLEKRTLRQSTNEKSLLSQSQLSPRL
KKKLRLNEDGSVDINGQQGEQGGQAQGAGGKKKRYIHDFLTQEQLLKEAVFTEFVNRKQL
EDLLRLEEEKKQFSLSKKFIDEGPKIIYKNSAKTDENLIIYPTQEVYDSTIGNYMQSFNQ
QKQIKEKKFRYHDPKTNKSYNTPEEFYKYRTILYKSEQNRYKYLIKVYKDLQEKKEELLE
EFEPKLKDKYNNNCNNSSDSDDSLSSSSQSSSNSATSSAGKKKKRKSQQKKKKQTKKAKS
QKSRISQTAAEREDDENDEENDDDNNNNQDDKIFIDQDSD
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences



