Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00421000Standard Name
RRM18 (RNA Recognition Motif-containing protein 18)Aliases
PreTt07225 | 46.m00280 | 3679.m00130Description
RRM18 RNA-binding family CELF1 RNA recognition motif 3 in CELF/bruno-like protein; RNA recognition motif-containing protein; 2014 reannotation incorporated RRM19 (TTHERM_00421010) into this gene model; RNA recognition motif domainGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- alpha6-beta1 integrin-CYR61 complex (IEA) | GO:0071116
- EGFR-Grb2-Sos complex (IEA) | GO:0070620
- external side of plasma membrane (IEA) | GO:0009897
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- obsolete ectoplasm (IEA) | GO:0043265
Molecular Function
- 3-hydroxybutyrate dehydrogenase activity (IEA) | GO:0003858
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- ABC-type maltose transporter activity (IEA) | GO:0015423
- ABC-type oligosaccharide transporter activity (IEA) | GO:0015422
- aminotriazole transporter activity (IEA) | GO:0015241
- Cys-tRNA(Pro) hydrolase activity (IEA) | GO:0043907
- ferric enterobactin:proton symporter activity (IEA) | GO:0015345
- high-affinity ferric iron transmembrane transporter activity (IEA) | GO:0015092
- lipopolysaccharide heptosyltransferase activity (IEA) | GO:0008920
- microtubule severing ATPase activity (IEA) | GO:0008568
- obsolete D-lysine oxidase activity (IEA) | GO:0043912
- obsolete myosin ATPase activity (IEA) | GO:0008570
- obsolete peroxisome-assembly ATPase activity (IEA) | GO:0008573
- obsolete phosphatidylinositol 3-kinase, class I, catalyst activity (IEA) | GO:0015072
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete toxin activity (IEA) | GO:0015070
- peptide-glutamate-alpha-N-acetyltransferase activity (IEA) | GO:1990190
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- prenyltransferase activity (IEA) | GO:0004659
- prephenate dehydrogenase (NADP+) activity (IEA) | GO:0004665
- proteinase activated receptor binding (IEA) | GO:0031871
- type 1 proteinase activated receptor binding (IEA) | GO:0031872
Biological Process
- cadmium ion transport (IEA) | GO:0015691
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- coenzyme A biosynthetic process (IEA) | GO:0015937
- external genitalia morphogenesis (IEA) | GO:0035261
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- glycolytic process from glycerol (IEA) | GO:0061613
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- laminaritriose transport (IEA) | GO:2001097
- long-term memory (IEA) | GO:0007616
- macrophage derived foam cell differentiation (IEA) | GO:0010742
- maltoheptaose metabolic process (IEA) | GO:2001122
- mating behavior (IEA) | GO:0007617
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of DNA amplification (IEA) | GO:1904524
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of peripheral B cell deletion (IEA) | GO:0002909
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete positive regulation of mitotic G1 cell cycle arrest in response to nitrogen starvation (IEA) | GO:1903694
- obsolete reproduction (IEA) | GO:0000003
- ommochrome biosynthetic process (IEA) | GO:0006727
- parathion metabolic process (IEA) | GO:0018952
- phagocytosis (IEA) | GO:0006909
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of response to water deprivation (IEA) | GO:1902584
- programmed cell death (IEA) | GO:0012501
- pronuclear fusion (IEA) | GO:0007344
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of macrophage derived foam cell differentiation (IEA) | GO:0010743
- regulation of phosphatidylcholine biosynthetic process (IEA) | GO:2001245
- regulation of satellite cell activation involved in skeletal muscle regeneration (IEA) | GO:0014717
- regulation of sprouting angiogenesis (IEA) | GO:1903670
- regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903689
- sesquiterpenoid biosynthetic process (IEA) | GO:0016106
- sodium-independent thromboxane transport (IEA) | GO:0071721
- sophorose transport (IEA) | GO:2001087
- sperm aster formation (IEA) | GO:0035044
- synaptic vesicle priming (IEA) | GO:0016082
- tetrasaccharide transport (IEA) | GO:2001098
- xanthophyll biosynthetic process (IEA) | GO:0016123
- xanthophyll catabolic process (IEA) | GO:0016124
- xylotriose transport (IEA) | GO:2001094
 Domains
Domains
- ( PF00076 ) RNA recognition motif
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
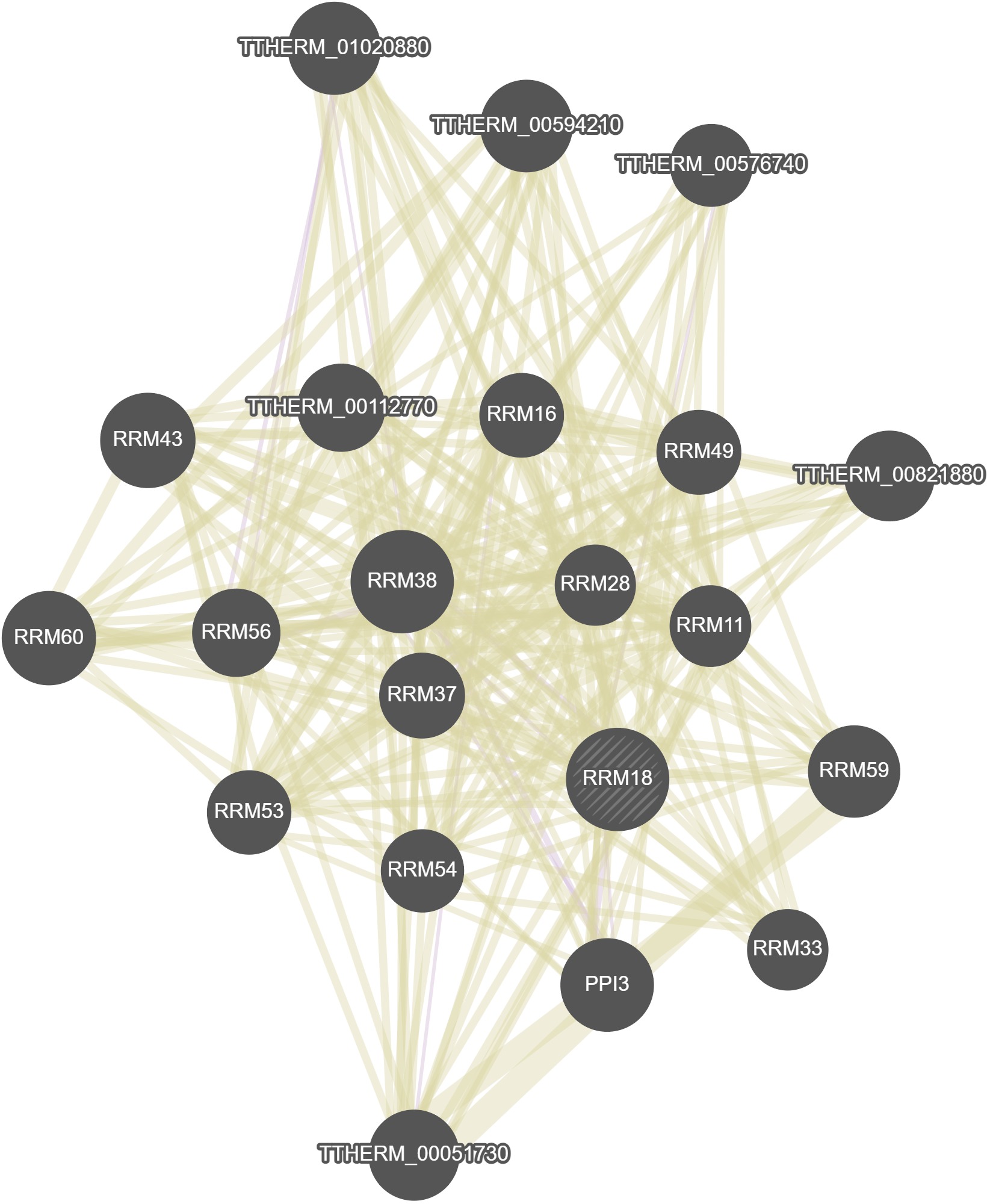
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00421000(coding)
ATGAATACCAACGAATACATAGACTAATTCGATTAAAATAAAAATATGGGAGCTTATTAC
GATAGCAATGGTTATGCTATGTAAAACTAGTAATAAATGGATTAAACCTACCAAAACCCT
CAAAACGAAATTAAGGAAGGTGGTGATCTACTTAAAATTAACAATACTTAAATTTTGATC
CGTAGATAGGAAAATACAGTCGATTTAAAACTCTTTGTCGGTTAACTACCAAAGACATGG
GAAAAAGAGCAAGTTAAAGATTTCTTTTCTAAATTCGGCAGAATTTATGAAGTTTAGATT
ATTAGAGACAATAAGGGATAACACAAGGGTTGCGCTTTTGTTAAATTTGCTTCAATGACT
GACGCAGAAAAAGCTATTGAAGCTGTAAAAAACACCACCTTTCCAGGAATGAAGAATAAC
GTTGAAATTAAATGGGCTGACAACGAAGAAGAAAGACTTGGTGTCAACTAAGACTCTGAC
CATAAATTATTTATTGGTTCTCTTCCTAAGTCTTGCACTGAATAAAATATTAAAGATATC
TTTGAATTTTTTGGTGAAATTGAAGAATTACACCTTATGAAGGATAACTAGTAAAACACA
AGATAAGCATTTTTAAAATTCAAACAAAAAGAAAAAGCTCACCTTGCTATCAGAAACTTA
AACTCTCAAGTCTACATCAATGATGCTGAAAATCCAATTGAAGTTAGATTCGCTAAGAAA
TATGTTAAGCAAGAAAAGCCAAATCCTACTCCAGCTCCTAAACCTGACCCTGCTCCTGCT
CCTTAGCAAATCAGCTGGTAAAAAGTATACTTCAAATACTACACTGAAGAGAATGAACCT
TACTACTTCCATCCTCTTCTAAATACTAGTGCCTTTGACAAACCTGGTCCTGGAAGCTTG
ATTTTAGATATTGATGGAAATCAAGAATACATAGATAACAACCCTAGACCTACAAGAAAC
GAACCTGTTGTTATTAATGATAGAAAGGATGTTTATGGCAAAAAGCATGGACCTCCTGGA
GCCAATCTTTTTATATTCCACTTGCCCAATGACTACAGAGATTCCGATTTAGAAAAACTC
TTTTCAGAATATGGTGAGGTAATTTCTGCTAGAGTCAATACAAGACCTGATGGAACAAGC
AAAGGATTTGGTTTCATTAGTTATAACCACCCATCTTAGGCAGAGGCTGCAATCACTGCT
CTCAACGGACTCTAGATTAAAAATAAGAGACTAAAAGTTGAAATAAAAAAAGAAGGAATG
GATAACAATGGAAACATGAAGCAATCCAACCCTGGATCTTTAAACCAAACTTCTTTCCAT
CCATTCTGA>TTHERM_00421000(gene)
CTTGGCTACCAGGCAAAAGAAAAATCATATAAATAATTAAAATTAGAAAGAGTTAATATC
AAAAATCTTATTATAAAGAGCAAGTTTTGAATATAAGTAAAACAGACATAAGAGATAAAA
TGAATACCAACGAATACATAGACTAATTCGATTAAAATAAAAATATGGGAGCTTATTACG
ATAGCAATGGTTATGCTATGTAAAACTAGTAATAAATGGATTAAACCTACCAAAACCCTC
AAAACGAAATTAAGGAAGGTGGTGATCTACTTAAAATTAACAATACTTAAATTTTGATCC
GTAGATAGGAAAATACAGTCGATTTAAAACTCTTTGTCGGTTAACTACCAAAGACATGGG
AAAAAGAGCAAGTTAAAGATTTCTTTTCTAAATTCGGCAGAATTTATGAAGTTTAGATTA
TTAGAGACAATAAGGGATAACACAAGGGTTGCGCTTTTGTTAAATTTGCTTCAATGACTG
ACGCAGAAAAAGCTATTGAAGCTGTAAAAAACACCACCTTTCCAGGAATGAAGAATAACG
TTGAAATTAAATGGGCTGACAACGAAGAAGAAAGACTTGGTGTCAACTAAGACTCTGACC
ATAAATTATTTATTGGTTCTCTTCCTAAGTCTTGCACTGAATAAAATATTAAAGATATCT
TTGAATTTTTTGGTGAAATTGAAGAATTACACCTTATGAAGGATAACTAGTAAAACACAA
GATAAGCATTTTTAAAATTCAAACAAAAAGAAAAAGCTCACCTTGCTATCAGAAACTTAA
ACTCTCAAGTCTACATCAATGATGCTGAAAATCCAATTGAAGTTAGATTCGCTAAGAAAT
ATGTTAAGCAAGAAAAGCCAAATCCTACTCCAGCTCCTAAACCTGACCCTGCTCCTGCTC
CTTAGCAAATCAGCTGGTAAAAAGTATACTTCAAATACTACACTGAAGAGAATGAACCTT
ACTACTTCCATCCTCTTCTAAATACTAGTGCCTTTGACAAACCTGGTCCTGGAAGCTTGA
TTTTAGATATTGATGGAAATCAAGAATACATAGATAACAACCCTAGACCTACAAGAAACG
AACCTGTTGTTATTAATGATAGAAAGGATGTTTATGGCAAAAAGCATGGACCTCCTGGAG
CCAATCTTTTTATATTCCACTTGCCCAATGACTACAGAGATTCCGATTTAGAAAAACTCT
TTTCAGAATATGGTGAGGTAATTTCTGCTAGAGTCAATACAAGACCTGATGGAACAAGCA
AAGGATTTGGTTTCATTAGTTATAACCACCCATCTTAGGCAGAGGCTGCAATCACTGCTC
TCAACGGACTCTAGATTAAAAATAAGAGACTAAAAGTTGAAATAAAAAAAGAAGGAATGG
ATAACAATGGAAACATGAAGCAATCCAACCCTGGATCTTTAAACCAAACTTCTTTCCATC
CATTCTGAATTATATTAAAGAAAGGACAATAAATAAAATAAAATTTATTTTTAATATCTT
AATTTAAGCTGAAAAATGAACAAAAAATAAAATAAATAAATAATTAAATAATTAAACAAA
TTATGTCTTCTAATTTTTTCTAAAAACCTTATTATATATTTTATTATTATATTATCTATG
CTTGCTTATTTCATTTATTATTTCTATAGCATCATGTATATAATTTTTTTTTACTTATTA
GTTTAATCGA>TTHERM_00421000(protein)
MNTNEYIDQFDQNKNMGAYYDSNGYAMQNQQQMDQTYQNPQNEIKEGGDLLKINNTQILI
RRQENTVDLKLFVGQLPKTWEKEQVKDFFSKFGRIYEVQIIRDNKGQHKGCAFVKFASMT
DAEKAIEAVKNTTFPGMKNNVEIKWADNEEERLGVNQDSDHKLFIGSLPKSCTEQNIKDI
FEFFGEIEELHLMKDNQQNTRQAFLKFKQKEKAHLAIRNLNSQVYINDAENPIEVRFAKK
YVKQEKPNPTPAPKPDPAPAPQQISWQKVYFKYYTEENEPYYFHPLLNTSAFDKPGPGSL
ILDIDGNQEYIDNNPRPTRNEPVVINDRKDVYGKKHGPPGANLFIFHLPNDYRDSDLEKL
FSEYGEVISARVNTRPDGTSKGFGFISYNHPSQAEAAITALNGLQIKNKRLKVEIKKEGM
DNNGNMKQSNPGSLNQTSFHPF