Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00433810Standard Name
HEN1 (HEN1 (Hua ENhancer) homolog 1)Aliases
PreTt26522 | 48.m00285 | 3831.m01622Description
HEN1 small RNA 2-prime-O-methyltransferase; RNA methyltransferase; stabilizes scnRNA by 2'-O-methylation; ensures DNA elimination in Tetrahymena; colocalizes and interacts with TWI1; expressed during growth and early conjugation; homolog of Arabidopsis HEN1; 3'-RNA ribose 2'-O-methyltransferase, Hen1Genome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- CURI complex (IEA) | GO:0032545
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- macronucleus (IDA) | GO:0031039
- terminal cisterna lumen (IEA) | GO:0014804
Molecular Function
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- deoxyribonucleoside binding (IEA) | GO:0032546
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- p53 binding (IEA) | GO:0002039
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- proteinase activated receptor binding (IEA) | GO:0031871
- RNA methyltransferase activity (IDA) | GO:0008173
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
Biological Process
- auxin metabolic process (IEA) | GO:0009850
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to diacyl bacterial lipopeptide (IEA) | GO:0071726
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- indoleacetic acid biosynthetic process via tryptophan (IEA) | GO:0009848
- microtubule-based peroxisome localization (IEA) | GO:0060152
- negative regulation of chemokine (C-C motif) ligand 1 production (IEA) | GO:0071653
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- ommochrome biosynthetic process (IEA) | GO:0006727
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of L-arginine import into cell (IEA) | GO:1902828
- positive regulation of response to water deprivation (IEA) | GO:1902584
- programmed DNA elimination (IDA) | GO:0031049
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- prothoracic disc morphogenesis (IEA) | GO:0007470
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of L-arginine import into cell (IEA) | GO:1902826
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of syncytium formation by plasma membrane fusion (IEA) | GO:0060142
- RNA catabolic process (IMP) | GO:0006401
- seed germination (IEA) | GO:0009845
- snRNA 2'-O-methylation (IDA) | GO:1990437
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- testosterone biosynthetic process (IEA) | GO:0061370
 Domains
Domains
No Data fetched for Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
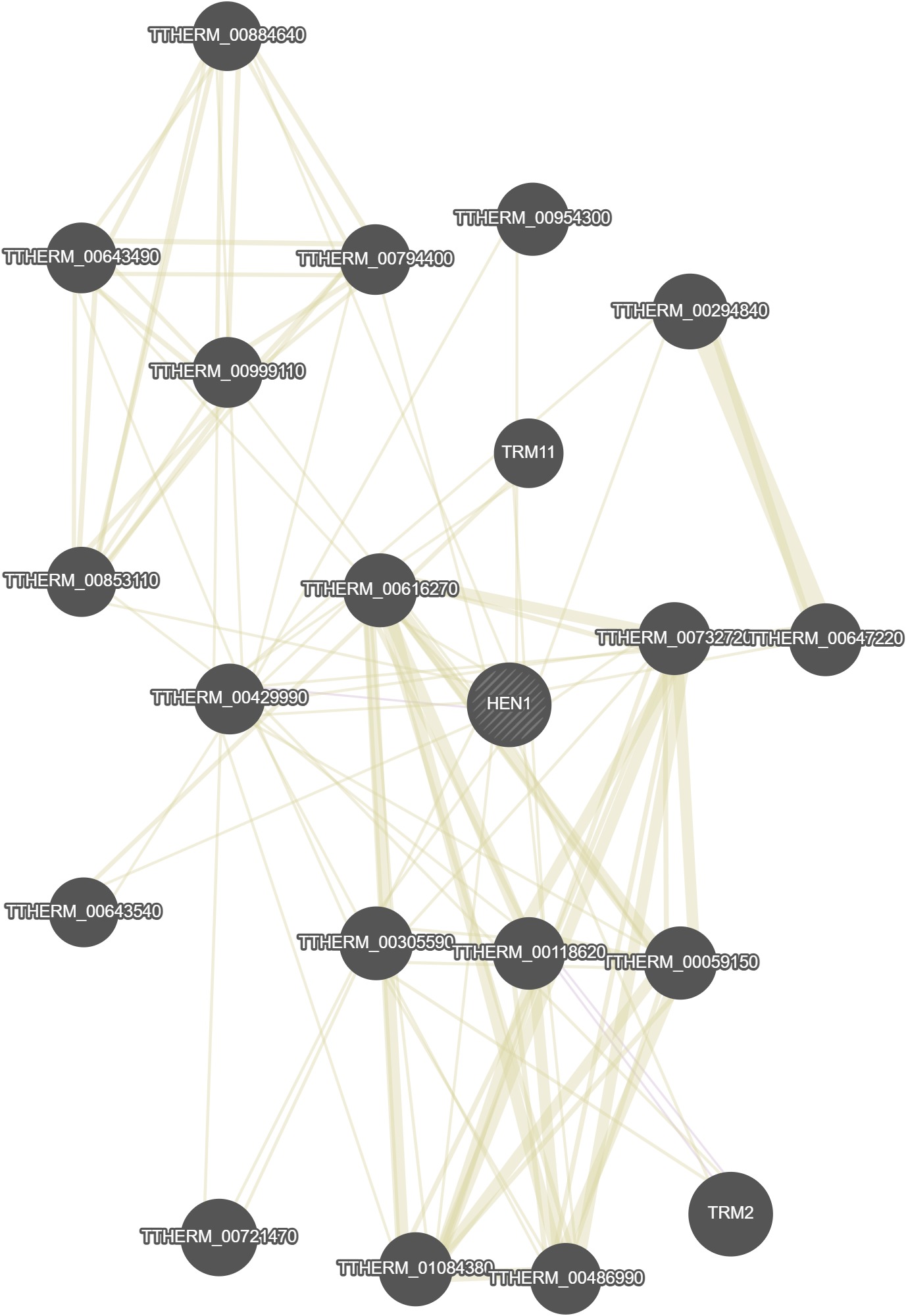
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No. Gene Name(s) Paragraph Text 2315 HEN1, TWI1 Twi1p-associated scnRNAs are 2'-O-methylated at their 3' ends by the RNA methyltransferase Hen1p (Kurth & Mochizuki 2009).
 Associated Literature
Associated Literature
- Ref:29021213: Noto T, Mochizuki K (2017) Whats, hows and whys of programmed DNA elimination in Tetrahymena. Open biology 7(10):
- Ref:19240163: Kurth HM, Mochizuki K (2009) 2'-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA (New York, N.Y.) 15(4):675-85
 Sequences
Sequences
>TTHERM_00433810(coding)
ATGATTGAAGCTTATGAAACAGATGTGTTTATGGATCCTATTGGTATGAAAGTTTGGGAA
AAAAGGCACTAATATGTAGCAACTAAGTTATCGGCTCTGAATTGTAAAAGAGTTTTGGAT
ATGGGAACAAATACTTGCAAGCTCATATAAAGACTTTCAAGGAGTTTGTAATTCACTTAA
ATAGATGGTTTAGATATCGATGGCTAATTGCTTGAGACTCAAGGAATCCAAAATGCGAAG
CCAGATTTGATATAAAATTAATATGCCAGTATGAGAGACCATTAGCTAGTGGTAAATCTA
TACCAAGGCAGTGCTTTGAATAAGATTTAACATTTGAAGGATTAGCAATACGATGCTGTA
ATATTAGTTGAACTCATTGAGCACTTATAAGTAGAAGATGTTTTTCTTATTGAGTAAAAT
TTGTTTGGCTTTTTGAGGCCACAATTTGTGATTGTGACTACACCTAACTCTGATTTTAAT
GTTTATTTTAACTTTAAAGAATAAGGAGTATTGTTTAGGGACAAAGATCACAAATTTGAG
TGGTCCTAAAACTAGTTTTAGATTTGGGCCTAAAAAGTATGCTAAAATTATGGATACAAA
GTAATAGAGTTGACAGGAGTAGGCGAACATAAAACAGAAGGAACTAAGAACGGTTTTTGT
ACACAAATAGTTGTATTTGAGAAAGATACCCAACAAGAGAAGTATCTGAACTTTGCTTTC
TTCAACCTTCAAGAGGGAGAAATTCGTTAAGTATGTTAAATTCTGTACCCATTTGAGAGT
AAAGAATAACATTTTCAAAGAGAAGTAGTAGACAGTATCAGATATATTCTGCATATTACT
GATAAGTAAAATTAATTCGAAGATGGTTCATACTAAAATTATACTACTCTCTCTCGCATA
ATGCAAAATCATTCAATATCAAGTAATTGGTAAATTTAAGGTGATTACTTTAAACTTAAA
ACCTACATCCAGAATATTTCTGAGTTTTTGGTACATGAAAATTAGTTCAATTTTCAAGAA
TCTTTCGTCACTCTCAATTATTAAGCCTAAATGGAAGATGAAGAAAATGAAGATTAGCTT
GAAAGCGATTCTGAAAATGTAAAGATGCAATAATAACAATATTACTTTTCTAACGATAAT
TGTTTCTCCACAAAAGATACCACTTATAGCAGTTTTTCCACTGCAGATAACTTATTCTCC
TAAAAAATTTAACTTGGTTAGCAATAAATGGCTTTAGAGGAAATAGAATTAGAAGATACA
ATTGATTATTGA>TTHERM_00433810(gene)
ATTAAAAGTGCAAAAAAAAAAAATTAGAAATCTACCAAATAAACAAAAAGAAAGAAAGAA
AAAAATTTATTTATATTAAAAATGATTGAAGCTTATGAAACAGATGTGTTTATGGATCCT
ATTGGTATGAAAGTTTGGGAAAAAAGGCACTAATATGTAGCAACTAAGTTATCGGCTCTG
AATTGTAAAAGAGTTTTGGATATGGGAACAAATACTTGCAAGCTCATATAAAGACTTTCA
AGGAGTTTGTAATTCACTTAAATAGATGGTTTAGATATCGATGGCTAATTGCTTGAGACT
CAAGGAATCCAAAATGCGAAGCCAGATTTGATATAAAATTAATATGCCAGTATGAGAGAC
CATTAGCTAGTGGTAAATCTATACCAAGGCAGTGCTTTGAATAAGATTTAACATTTGAAG
GATTAGCAATACGATGCTGTAATATTAGTTGAACTCATTGAGCACTTATAAGTAGAAGAT
GTTTTTCTTATTGAGTAAAATTTGTTTGGCTTTTTGAGGCCACAATTTGTGATTGTGACT
ACACCTAACTCTGATTTTAATGTTTATTTTAACTTTAAAGAATAAGGAGTATTGTTTAGG
GACAAAGATCACAAATTTGAGTGGTCCTAAAACTAGTTTTAGATTTGGGCCTAAAAAGTA
TGCTAAAATTATGGATACAAAGTAATAGAGTTGACAGGAGTAGGCGAACATAAAACAGAA
GGAACTAAGAACGGTTTTTGTACACAAATAGTTGTATTTGAGAAAGATACCCAACAAGAG
AAGTATCTGAACTTTGCTTTCTTCAACCTTCAAGAGGGAGAAATTCGTTAAGTATGTTAA
ATTCTGTACCCATTTGAGAGTAAAGAATAACATTTTCAAAGAGAAGTAGTAGACAGTATC
AGATATATTCTGCATATTACTGATAAGTAAAATTAATTCGAAGATGGTTCATACTAAAAT
TATACTACTCTCTCTCGCATAATGCAAAATCATTCAATATCAAGTAATTGGTAAATTTAA
GGTGATTACTTTAAACTTAAAACCTACATCCAGAATATTTCTGAGTTTTTGGTACATGAA
AATTAGTTCAATTTTCAAGAATCTTTCGTCACTCTCAATTATTAAGCCTAAATGGAAGAT
GAAGAAAATGAAGATTAGCTTGAAAGCGATTCTGAAAATGTAAAGATGCAATAATAACAA
TATTACTTTTCTAACGATAATTGTTTCTCCACAAAAGATACCACTTATAGCAGTTTTTCC
ACTGCAGATAACTTATTCTCCTAAAAAATTTAACTTGGTTAGCAATAAATGGCTTTAGAG
GAAATAGAATTAGAAGATACAATTGATTATTGAAGCATTACAAAATAAATGGAAAAAAAT
TAAAAATACTTCCTTCATTTAATTAACTAAAAACAATAAAATCTTAAAATAATTAATATG
TTTAAATTTATCCAAAACCTTGATAAATAATTTCAAAGACTTCACTTTTAACTCTATCTT
TAAAAATAATAAATTAATCATATTCATAAGCTGTAGATTTTATACTATTTATTCGTATT>TTHERM_00433810(protein)
MIEAYETDVFMDPIGMKVWEKRHQYVATKLSALNCKRVLDMGTNTCKLIQRLSRSLQFTQ
IDGLDIDGQLLETQGIQNAKPDLIQNQYASMRDHQLVVNLYQGSALNKIQHLKDQQYDAV
ILVELIEHLQVEDVFLIEQNLFGFLRPQFVIVTTPNSDFNVYFNFKEQGVLFRDKDHKFE
WSQNQFQIWAQKVCQNYGYKVIELTGVGEHKTEGTKNGFCTQIVVFEKDTQQEKYLNFAF
FNLQEGEIRQVCQILYPFESKEQHFQREVVDSIRYILHITDKQNQFEDGSYQNYTTLSRI
MQNHSISSNWQIQGDYFKLKTYIQNISEFLVHENQFNFQESFVTLNYQAQMEDEENEDQL
ESDSENVKMQQQQYYFSNDNCFSTKDTTYSSFSTADNLFSQKIQLGQQQMALEEIELEDT
IDY