Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00446570Standard Name
HAP5 (Heme Activator Protein)Aliases
PreTt03924 | 51.m00290 | 3704.m00146Description
HAP5 histone-like transcription factor and archaeal histone protein; Transcription and DNA Replication RegulatorsGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- EGFR-Grb2-Sos complex (IEA) | GO:0070620
- ionotropic glutamate receptor complex (IEA) | GO:0008328
Molecular Function
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- CAP-Gly domain binding (IEA) | GO:0071794
- lanosterol synthase activity (IEA) | GO:0000250
- microtubule severing ATPase activity (IEA) | GO:0008568
- mitochondrial single subunit type RNA polymerase activity (IEA) | GO:0001065
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase (ATP) activity (IEA) | GO:0004612
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
Biological Process
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to dopamine (IEA) | GO:1903351
- cornification (IEA) | GO:0070268
- endoderm development (IEA) | GO:0007492
- ergosteryl 3-beta-D-glucoside biosynthetic process (IEA) | GO:1904463
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- inactivation of recombination (HML) (IEA) | GO:0007537
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete extracellular carbohydrate transport (IEA) | GO:0006859
- obsolete maintenance of dosage compensation (IEA) | GO:0007551
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- ommochrome biosynthetic process (IEA) | GO:0006727
- photoreceptor cell morphogenesis (IEA) | GO:0008594
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of dendrite extension (IEA) | GO:1903861
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of stomatal opening (IEA) | GO:1902458
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- prothoracic disc morphogenesis (IEA) | GO:0007470
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of plant epidermal cell differentiation (IEA) | GO:1903888
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- transcription by RNA polymerase V (IEA) | GO:0001060
- verruculogen metabolic process (IEA) | GO:1902179
- vestibular receptor cell differentiation (IEA) | GO:0060114
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
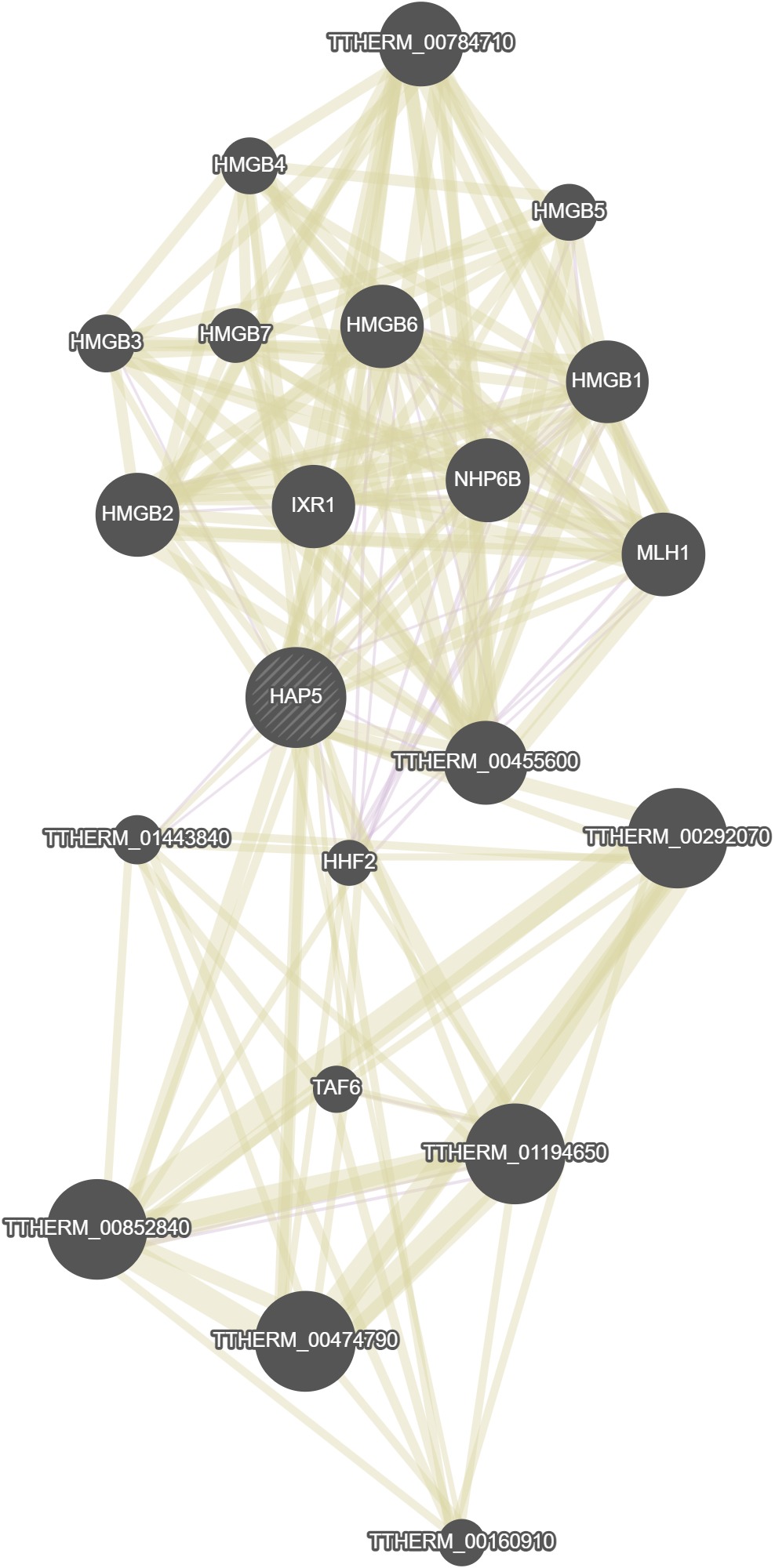
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:31932604: Nabeel-Shah S, Ashraf K, Saettone A, Garg J, Derynck J, Lambert JP, Pearlman RE, Fillingham J (2020) Nucleus-specific linker histones Hho1 and Mlh1 form distinct protein interactions during growth, starvation and development in Tetrahymena thermophila. Scientific reports 10(1):168
 Sequences
Sequences
>TTHERM_00446570(coding)
ATGGATTAAGAATAGTTGGAATTCGATTATGAAGATGAGTTTCAGGATCTAAACTTAGAG
TAGTAGCTAGCATAGCCTACAAGGTTTTTAAAAAGCTAAAATGGTGAAAAACAATAAATA
TAAACTAATACAGAAATGTAGAATAAAGAAGATATTACTAATTAAAAGAAGCATTTATAA
AAGCCTAAAAAGCCTTTGAGTGCATACTTTTTATTTATTTAAGACTATAAGCAAAAATGT
GGAAGCAAAATAAACTTGAAAGAAATGGGAAATATATGGAATGAAACTTCGTAAGATGAA
AAGCAAATATACTATAAAAAATAAGAAGAAGAAAAAGCTCGCTTTAAAAAAGAAATGGAC
GAATATATTGAATAGCAAGGTGACGAGGAAAATGAAGAACAATTAGATAATGAATTAGAT
ATTGCTTTTCCTATATCAAGGATTAAAGGTATTCTTAAAGTTGATGAAGACAATAAACTA
TTAAGCAAAGACACACTCCTTTACTTTGAAAAAGCTACAGAATTATTTGGGGGTTATTTA
GCTTAAAAATGTTTTAGAAATATGCAATAAAATCGCAGAAAAAAATTATAAACTGGTGAT
CTATATGAAGTAATTAAAAGGGACATGTACTTGAACTTCTTATAATGTATGGTGCCTTAA
GAAGCAACTGGTGAAGAAGAAAACCAATAAAATGTAATTAGTGGAAGCTAGCAGTCATCA
AAAACAAAATCTCGCAAAAGATTAGTCATTGCTGACGATGAAGATGTAAACTAAGAAGGC
AACATGGAAATAGAAAATAAAGATTTAGAGTAATAGGAGATTCCTGAATCAAGTAATAAA
AAAAATGTTAATACTAAGAGCAATAATAAACCAGGTAAAAAAGAAAAGGTTGTGCTACCT
CCTGCTAATAATAGCAGGATCGACAGCTTTTTTACTAAAAAGTGA>TTHERM_00446570(gene)
AAATCTTTTGGATATAAATTAAATTTTTGTAAATTTACATTTAATAGTAAACAAATTTTA
TAAGATATTTTAAGACGATCAAATGGATTAAGAATAGTTGGAATTCGATTATGAAGATGA
GTTTCAGGATCTAAACTTAGAGTAGTAGCTAGCATAGCCTACAAGGTTTTTAAAAAGCTA
AAATGGTGAAAAACAATAAATATAAACTAATACAGAAATGTAGAATAAAGAAGATATTAC
TAATTAAAAGAAGCATTTATAAAAGCCTAAAAAGCCTTTGAGTGCATACTTTTTATTTAT
TTAAGACTATAAGCAAAAATGTGGAAGCAAAATAAACTTGAAAGAAATGGGAAATATATG
GAATGAAACTTCGTAAGATGAAAAGCAAATATACTATAAAAAATAAGAAGAAGAAAAAGC
TCGCTTTAAAAAAGAAATGGACGAATATATTGAATAGCAAGGTGACGAGGAAAATGAAGA
ACAATTAGATAATGAATTAGATATTGCTTTTCCTATATCAAGGATTAAAGGTATTCTTAA
AGTTGATGAAGACAATAAACTATTAAGCAAAGACACACTCCTTTACTTTGAAAAAGCTAC
AGAATTATTTGGGGGTTATTTAGCTTAAAAATGTTTTAGAAATATGCAATAAAATCGCAG
AAAAAAATTATAAACTGGTGATCTATATGAAGTAATTAAAAGGGACATGTACTTGAACTT
CTTATAATGTATGGTGCCTTAAGAAGCAACTGGTGAAGAAGAAAACCAATAAAATGTAAT
TAGTGGAAGCTAGCAGTCATCAAAAACAAAATCTCGCAAAAGATTAGTCATTGCTGACGA
TGAAGATGTAAACTAAGAAGGCAACATGGAAATAGAAAATAAAGATTTAGAGTAATAGGA
GATTCCTGAATCAAGTAATAAAAAAAATGTTAATACTAAGAGCAATAATAAACCAGGTAA
AAAAGAAAAGGTTGTGCTACCTCCTGCTAATAATAGCAGGATCGACAGCTTTTTTACTAA
AAAGTGAATATTTTTATTCTCATCCGAAATAAAAAAGTTAATTTAATTTAATAGTAAACA
AATATTTGTATTAAGTATAAATAATTATTTTGAAAAATTA>TTHERM_00446570(protein)
MDQEQLEFDYEDEFQDLNLEQQLAQPTRFLKSQNGEKQQIQTNTEMQNKEDITNQKKHLQ
KPKKPLSAYFLFIQDYKQKCGSKINLKEMGNIWNETSQDEKQIYYKKQEEEKARFKKEMD
EYIEQQGDEENEEQLDNELDIAFPISRIKGILKVDEDNKLLSKDTLLYFEKATELFGGYL
AQKCFRNMQQNRRKKLQTGDLYEVIKRDMYLNFLQCMVPQEATGEEENQQNVISGSQQSS
KTKSRKRLVIADDEDVNQEGNMEIENKDLEQQEIPESSNKKNVNTKSNNKPGKKEKVVLP
PANNSRIDSFFTKK