Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00459230Standard Name
DMC1 (Disrupted Meiotic CDNA 1)Aliases
PreTt20354 | 53.m00256 | 3712.m00114Description
DMC1 meiotic recombination protein DMC1 putative; Meiosis-specific RecA; homolog of S. cerevisiae DMC1; DNA recombination and repair protein Rad51-like, C-terminalGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- chloroplast ribosome (IEA) | GO:0043253
- EGFR-Grb2-Sos complex (IEA) | GO:0070620
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- micronucleus (IDA) | GO:0031040
- obsolete ectoplasm (IEA) | GO:0043265
- obsolete integral component of mitochondrial membrane (IEA) | GO:0032592
- phosphatidylinositol transporter complex (IEA) | GO:1902556
- superoxide dismutase complex (IEA) | GO:1902693
Molecular Function
- [citrate (pro-3S)-lyase] ligase activity (IEA) | GO:0008771
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-octaprenyl-6-methoxyphenol hydroxylase activity (IEA) | GO:0008681
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- alpha-humulene synthase activity (IEA) | GO:0080017
- bacterial-type RNA polymerase core enzyme binding (IEA) | GO:0001000
- ferric triacetylfusarinine C:proton symporter activity (IEA) | GO:0015346
- G protein-coupled neurotensin receptor activity (IEA) | GO:0016492
- lanosterol synthase activity (IEA) | GO:0000250
- microtubule severing ATPase activity (IEA) | GO:0008568
- mitochondrial RNA polymerase binding promoter specificity activity (IEA) | GO:0000998
- mitochondrial RNA polymerase core promoter sequence-specific DNA binding (IEA) | GO:0000997
- mitochondrial single subunit type RNA polymerase activity (IEA) | GO:0001065
- MutSbeta complex binding (IEA) | GO:0032408
- obsolete calcium-dependent secreted phospholipase A2 activity (IEA) | GO:0004625
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete phosphatidylinositol 3-kinase, class I, catalyst activity (IEA) | GO:0015072
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylinositol-3-phosphate binding (IEA) | GO:0032266
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase (ATP) activity (IEA) | GO:0004612
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- phosphopantothenate--cysteine ligase activity (IEA) | GO:0004632
- Pup ligase activity (IEA) | GO:0061664
- pyrimidine nucleotide-sugar transmembrane transporter activity (IEA) | GO:0015165
- regulatory region RNA binding (IEA) | GO:0001069
- ribonuclease MRP activity (IEA) | GO:0000171
- RNA polymerase III activity (IEA) | GO:0001056
- RNA-binding transcription regulator activity (IEA) | GO:0001070
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly (IEA) | GO:0001075
- transcription factor activity, RNA polymerase III type 1 promoter sequence-specific binding, TFIIIB recruiting (IEA) | GO:0001005
- transcription regulatory region RNA binding (IEA) | GO:0001068
- tRNA threonylcarbamoyladenosine dehydratase (IEA) | GO:0061503
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
- UDP-galactopyranose mutase activity (IEA) | GO:0008767
- UFM1 ligase activity (IEA) | GO:0061666
Biological Process
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- absorption of visible light (IEA) | GO:0016038
- adult visceral muscle development (IEA) | GO:0007524
- alkaloid biosynthetic process (IEA) | GO:0009821
- basement membrane organization (IEA) | GO:0071711
- cell surface receptor signaling pathway involved in heart development (IEA) | GO:0061311
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular component organization (IEA) | GO:0016043
- cellular response to carbendazim (IEA) | GO:0072762
- cellular response to dopamine (IEA) | GO:1903351
- cellular response to hydroxyurea (IEA) | GO:0072711
- cytosolic calcium ion transport (IEA) | GO:0060401
- di-, tri-valent inorganic cation transport (IEA) | GO:0015674
- double-strand break repair (IMP) | GO:0006302
- endoderm development (IEA) | GO:0007492
- endodermal cell fate determination (IEA) | GO:0007493
- entry into host via enzymatic degradation of host cuticle (IEA) | GO:0085028
- epithelium development (IEA) | GO:0060429
- establishment or maintenance of polarity of embryonic epithelium (IEA) | GO:0016332
- evoked neurotransmitter secretion (IEA) | GO:0061670
- garland cell differentiation (IEA) | GO:0007514
- glycolytic process from mannose through fructose-6-phosphate (IEA) | GO:0061619
- hair cycle process (IEA) | GO:0022405
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- homogentisate metabolic process (IEA) | GO:1901999
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- interleukin-19 production (IEA) | GO:0032622
- interleukin-2 production (IEA) | GO:0032623
- intraciliary retrograde transport (IEA) | GO:0035721
- laminaritriose transport (IEA) | GO:2001097
- leg disc proximal/distal pattern formation (IEA) | GO:0007479
- lung connective tissue development (IEA) | GO:0060427
- malonic acid transmembrane transport (IEA) | GO:1901553
- mannosyl-inositol phosphorylceramide metabolic process (IEA) | GO:0006675
- meiotic nuclear division (IMP) | GO:0007126
- micromitophagy (IEA) | GO:0000424
- mitochondrial membrane organization (IEA) | GO:0007006
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of chemokine (C-C motif) ligand 20 production (IEA) | GO:1903885
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of interleukin-1 production (IEA) | GO:0032692
- negative regulation of interleukin-4-mediated signaling pathway (IEA) | GO:1902215
- negative regulation of iron ion import across plasma membrane (IEA) | GO:1904439
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- neutral lipid metabolic process (IEA) | GO:0006638
- nitrate transmembrane transport (IEA) | GO:0015706
- Notch signaling pathway involved in negative regulation of venous endothelial cell fate commitment (IEA) | GO:2000796
- obsolete activation of MAPK activity (IEA) | GO:0000187
- obsolete extracellular carbohydrate transport (IEA) | GO:0006859
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete multi-organism cellular localization (IEA) | GO:1902581
- obsolete negative regulation of adhesion of symbiont to host epithelial cell (IEA) | GO:1905227
- obsolete negative regulation of cytochrome-c oxidase activity (IEA) | GO:1905376
- obsolete negative regulation of histone H3-K18 acetylation (IEA) | GO:1902030
- obsolete negative regulation of recombination within rDNA repeats (IEA) | GO:0000020
- obsolete perturbation of host protein kinase-mediated signal transduction (IEA) | GO:0075130
- obsolete positive regulation of G-protein gamma subunit-mediated signal transduction in response to host (IEA) | GO:0075166
- obsolete primary charge separation in photosystem I (IEA) | GO:0009770
- obsolete prothoracic morphogenesis (IEA) | GO:0007471
- obsolete regulation of apurinic/apyrimidinic endodeoxyribonuclease activity (IEA) | GO:0052721
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete reproduction (IEA) | GO:0000003
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- obsolete sodium/potassium transport (IEA) | GO:0006815
- ommochrome biosynthetic process (IEA) | GO:0006727
- pachytene (IEA) | GO:0000239
- phagocytosis (IEA) | GO:0006909
- phagocytosis, engulfment (IEA) | GO:0006911
- phenylpropanoid metabolic process (IEA) | GO:0009698
- photoreceptor cell morphogenesis (IEA) | GO:0008594
- plant-type cell wall modification (IEA) | GO:0009827
- polyamine acetylation (IEA) | GO:0032917
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of dendrite extension (IEA) | GO:1903861
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of inner ear receptor cell differentiation (IEA) | GO:2000982
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of response to water deprivation (IEA) | GO:1902584
- programmed cell death (IEA) | GO:0012501
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- prothoracic disc morphogenesis (IEA) | GO:0007470
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of chitin metabolic process (IEA) | GO:0032882
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of plant epidermal cell differentiation (IEA) | GO:1903888
- regulation of protein localization to cell division site (IEA) | GO:1901900
- regulation of thyroid gland epithelial cell proliferation (IEA) | GO:1904441
- regulation of transporter activity (IEA) | GO:0032409
- response to singlet oxygen (IEA) | GO:0000304
- RNA polymerase III type 1 promoter transcriptional preinitiation complex assembly (IEA) | GO:0000999
- single-organism intracellular transport (IEA) | GO:1902582
- sperm aster formation (IEA) | GO:0035044
- sperm DNA decondensation (IEA) | GO:0035041
- striated muscle myosin thick filament assembly (IEA) | GO:0071688
- synaptic vesicle docking (IEA) | GO:0016081
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- transcription by RNA polymerase IV (IEA) | GO:0001059
- transcription by RNA polymerase V (IEA) | GO:0001060
- vesicle coating (IEA) | GO:0006901
- visceral muscle development (IEA) | GO:0007522
- wing disc morphogenesis (IEA) | GO:0007472
 Domains
Domains
- ( PF08423 ) Rad51
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
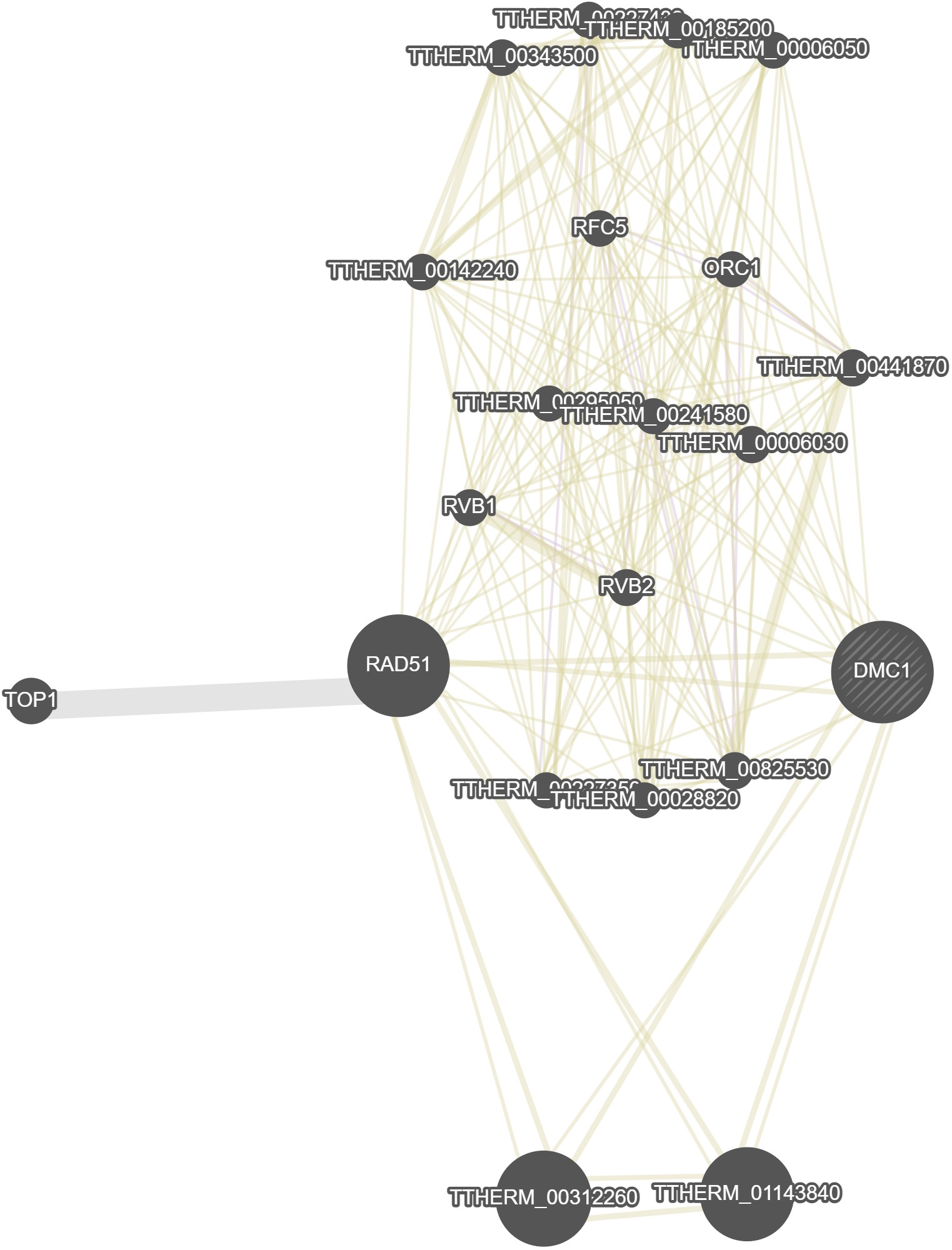
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:21483758: Howard-Till RA, Lukaszewicz A, Loidl J (2011) The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. PLoS genetics 7(3):e1001359
 Sequences
Sequences
>TTHERM_00459230(coding)
ATGAACGATTGTTACTCTGATAAAGAAGATAATGAAGAATAAAACTAAATTGCTCAAGAA
GAGATCTTTTTAGTAGAAATGCTTGCAACTGAGGGAGTTAACAATAATGAAATACAAAAG
CTTAAAAAGAATGGAATCCTTTCACTCAAATCTTTAGTTATGAACACAAAGAGGGATCTT
GTTAATATCTATGGAATTCCTGATAATAAAGCTGATTCATATGTCAAAAAAGCTTCAGAA
ATATTAGCTCGATCTGAAAACAGCAGGTTATTCAGCAGTGAGTTTGTTCTCGGTACTACT
GTACTATAAAGAAGGAGTTAAATAAGAAGAATATCAACAGGATCCAAGGCTTTAGATGAT
ATATTGAATGGGGGAATAGAGTCTCAAAGCATAACAGAATTTTATGGAGAATATCGTTCA
GGCAAAACATAAATAGCTCATACTGCATGCGTTCTGGCTCAATCTTAAGATCATTGCTAA
AGTCCAGGAAAAGTTTTATACATTGATACTGAAGGAACCTTCAGACCTGAAAGAATATGC
TAAATAGCTTCGCACTACGGAATGGAGGGTGAATATGCCCTAAGTAATATAATTTATGGG
AGAGCCTATAACGTAGATTAATAAAACACATTGCTTATTAAAGGAGCCCAATTAATGGTA
GAGGAAAATTGTTTTGCTCTACTAGTTGTTGATTCAATAATGGCTAATTTTAGATGCGAT
TTTTCTGGAAGAGGAGATCTCTCTGAAAGACAATAAGCACTTGGCAAGTTTATGAGCAGA
TTATAAAGAATGGCTGCTGAATTTAATATCGCAGTGATTATAACAAACTAAGTTATGGCT
GATCCAAGTGGAGCCATGACTGGAGGCGCTATACCCTAGCCCAAGCCAATTGGAGGCCAT
ATTTTAGCCCATGCATCGACATAGCGCTTATTCATGAAGAAGAAAACTGATAACATCCGT
AAAGTAAAGCTGGTTGATTCACCTTATTTGTAAGATAAAGAAGTTGATATTATGGTTAGC
GACAGAGGAGTAGGAGATGTTGAATGTGATAAAAAGCCTTCTACTGGTTGA>TTHERM_00459230(gene)
TTTCTCCCAATTTTACTTTTAATTATTTTTTCACTCATTTACAGTTGCTTATGTAAAATT
TAACAAACTAAAAAACAAGAAGAATCTAACTAGCTGCACTGAAAAGCACTACAAAAAATT
TTTTTTATTTTCTTTATAATTCTACAATTTTCATTCAATGTATATAAAAAATACTAAACC
CAATCTTTTACAAACTCAAAAAAAAATATTTTTATCATAAATATTCTATTTACATTTTAA
GCAAATACCATCTTTAGGAAAATGAACGATTGTTACTCTGATAAAGAAGATAATGAAGAA
TAAAACTAAATTGCTCAAGAAGAGATCTTTTTAGTAGAAATGCTTGCAACTGAGGGAGTT
AACAATAATGAAATACAAAAGCTTAAAAAGAATGGAATCCTTTCACTCAAATCTTTAGTT
ATGAACACAAAGAGGGATCTTGTTAATATCTATGGAATTCCTGATAATAAAGCTGATTCA
TATGTCAAAAAAGCTTCAGAAATATTAGCTCGATCTGAAAACAGCAGGTTATTCAGCAGT
GAGTTTGTTCTCGGTACTACTGTACTATAAAGAAGGAGTTAAATAAGAAGAATATCAACA
GGATCCAAGGCTTTAGATGATATATTGAATGGGGGAATAGAGTCTCAAAGCATAACAGAA
TTTTATGGAGAATATCGTTCAGGCAAAACATAAATAGCTCATACTGCATGCGTTCTGGCT
CAATCTTAAGATCATTGCTAAAGTCCAGGAAAAGTTTTATACATTGATACTGAAGGAACC
TTCAGACCTGAAAGAATATGCTAAATAGCTTCGCACTACGGAATGGAGGGTGAATATGCC
CTAAGTAATATAATTTATGGGAGAGCCTATAACGTAGATTAATAAAACACATTGCTTATT
AAAGGAGCCCAATTAATGGTAGAGGAAAATTGTTTTGCTCTACTAGTTGTTGATTCAATA
ATGGCTAATTTTAGATGCGATTTTTCTGGAAGAGGAGATCTCTCTGAAAGACAATAAGCA
CTTGGCAAGTTTATGAGCAGATTATAAAGAATGGCTGCTGAATTTAATATCGCAGTGATT
ATAACAAACTAAGTTATGGCTGATCCAAGTGGAGCCATGACTGGAGGCGCTATACCCTAG
CCCAAGCCAATTGGAGGCCATATTTTAGCCCATGCATCGACATAGCGCTTATTCATGAAG
AAGAAAACTGATAACATCCGTAAAGTAAAGCTGGTTGATTCACCTTATTTGTAAGATAAA
GAAGTTGATATTATGGTTAGCGACAGAGGAGTAGGAGATGTTGAATGTGATAAAAAGCCT
TCTACTGGTTGATTTTATAAAAAATAGAGTAAAGAATATTCTTTGAGAAAGTTAGTTAAA
TGAATAAAAAATAATATAAATAAATGGTTTATAAGATGTATTATACAAGTTTTATTAAAT
TATAATTGTAGT>TTHERM_00459230(protein)
MNDCYSDKEDNEEQNQIAQEEIFLVEMLATEGVNNNEIQKLKKNGILSLKSLVMNTKRDL
VNIYGIPDNKADSYVKKASEILARSENSRLFSSEFVLGTTVLQRRSQIRRISTGSKALDD
ILNGGIESQSITEFYGEYRSGKTQIAHTACVLAQSQDHCQSPGKVLYIDTEGTFRPERIC
QIASHYGMEGEYALSNIIYGRAYNVDQQNTLLIKGAQLMVEENCFALLVVDSIMANFRCD
FSGRGDLSERQQALGKFMSRLQRMAAEFNIAVIITNQVMADPSGAMTGGAIPQPKPIGGH
ILAHASTQRLFMKKKTDNIRKVKLVDSPYLQDKEVDIMVSDRGVGDVECDKKPSTG