Gene Model Identifier
TTHERM_00460480
Standard Name
COM1
(Meiotic double-strand break-induced reCOMbination 1)
Aliases
PreTt25699 | 54.m00155 | 3823.m01074
Description
COM1 Com1p is required for meiotic double-strand break repair and full homologous pairing; homolog of S. cerevisiae COM1/SAE2; hypothetical protein
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
>TTHERM_00460480(coding)
ATGCTTGGGTTAGAATAAAATTGTTTTATTAAAGAATACAGGAAGTAGGTAGATAAAGAG
GCTCTAAATTAAATATAAGATGCTGCTTTATCAAATTTAATGAATGCATTTGGGTAGATG
AATTAAACATATTCTGTACTTTTCAAAGATATTTGTCTTACTTTTGGCGCTTTATTATCA
ATAAAGGAAGAGTTAAAGGACTATCGCTAGGAAATGTTTTAGAAATAAAGCGAGAAAATT
ATCTAGGAGAAGATTCAGAAGGAAATGATTCAATAAAAAGATGAGCAATTGGAATTCCTC
TAAAAGTAATTGAAAATAGTTTAACTGCAATCTGATTAAGATAGAAATTCTAAACAGGCA
ATTCAAGAGAGGGATTAGGCCATAAGTTCATATAAATAAAGACTTTTTGATTTATAAGAA
AATGAAAAGAAATTGCTCAGCGAAAAGTCTTAGCTTCAGACTAGAATTGAGTTACTTGAA
GAATAATAAAATTCTAACAAAGTTGCAATCTTAGAAAAGCAAACCACTCTTCATTAAGGT
TATAAACTCACATTTTTAAATAACGAATTAAAAGAATCTCTTATTGAACAACCTTCAAAT
GTATTTTAATAAGAAAACGGGGTTAATAAAAATAAAATATATAGCTTGGATAATAATTAG
ATATAAAAGAATAATACTTCCCTTTTAAGTAAATTTAAGTAAGACCAATCTGATTAAGAG
GTGTAAAATAATACATAACTTGCTCAAAAAAACTAATAGGATATTTATTATAGTAACAAA
ATAAATCAGATAAAACCTGAAACATTTTCAAATCAAGATCCTTCAGTTTATTTACAAGAA
AGCTATGTATAAAGTTCACTTAATGCAAATAATATTTTAGGTTCAAAAAAGATTACAGAT
GAAGAGATTTTTATGATGTAAAAAAGAGAAGCTGAAATTAAAAAAAAGAAATAGGAAGAT
GAACATTTAAAGAATTAATAATACATGTCAAGTTTGAATTACAATAAAATAATTTCTGAT
AAAAATAAAAATTAGAATTTAATGAATAATAAAATTATTGAGTAATCTAAAGAGAGTTCT
ATGGGATCTTTATTCAATAAGACAGCTTTTGATCAAAGCTCAATTAATAATCAAAGTAAT
GCTAGTGTAAGAATGGAATTAGAAGATTCTAGTGCTGGATAACAGCTTTAAAGATATAAA
GTAAGCTTAAATGATAGAAAAATGGTAGCTGAAAATAATCTAAATGCAACAGATGATTTA
TTAAAAAATAAAATGGAGCAGGAAGATAATGAAAAAGAGAGTTCTGACTCTTAGTATAAC
TGGAATTTAAAAATCGATTTGAAAAACAATAACGAATAATAGCAGCAATTTCCATAGGAA
AGCAATAACAATAGTGGAAACAAAAAGAGATTCAAAAAGTAACAATTAAAAAAATCAGAT
TCTTTTGCCACAAATGCAAACTAGAATTCTGAATTAGAAAACACCACAGCTAGTGTTATT
AGTTTACAAAACTAACAATAAAATTAAAATTATGAAATTTAAGATCAATAAAGCAATTAA
ATGCTCTTAGGTCAAAAGAAAAATTTAAATATTAATAAAGGTAGAGTTGCTAATAATAAA
TTTATGAAAGACATAGAATTAGATGATGAAAGTGGGGACAAAGACTAACCCTAATAAAAT
GATATATTTGCTCAGTTTCAATATAAAAAAAGAGGTACTTCAAACTAAGAAGTTCAAGTA
GCTGAAACTTTTAACATGTATAACAAATAAAATTAGAGACCATTAAATCCTCGTAAAAAA
GAAGATAGAGAAAAATTAAAAACTTTTAATTGTGGCAGATGCAAAGCCGCATATCAAACT
TTGGGAGAAGAATTCTGTGGTAATTGTTCTAAACACAGATCAGAATACACGCCTCCTCAC
ACTCCCGAAGCCCATTATTAAAAATTTTCCTAGTCAGAAGATGATTCTAATTGA
>TTHERM_00460480(gene)
ATCTGCATTTTAAATACATAAGAATTCATTAACTAACAAATATACCAAGAATCTCTCATG
TAAATTACTAATTTGATATTTATCTCTCAGTATAAACAAACTTTAAAATTATAATTGTAT
AGTGGTATAAATAATAAATATAAATTTTTGAGCAAAAATGCTTGGGTTAGAATAAAATTG
TTTTATTAAAGAATACAGGAAGTAGGTAGATAAAGAGGCTCTAAATTAAATATAAGATGC
TGCTTTATCAAATTTAATGAATGCATTTGGGTAGATGAATTAAACATATTCTGTACTTTT
CAAAGATATTTGTCTTACTTTTGGCGCTTTATTATCAATAAAGGAAGAGTTAAAGGACTA
TCGCTAGGAAATGTTTTAGAAATAAAGCGAGAAAATTATCTAGGAGAAGATTCAGAAGGA
AATGATTCAATAAAAAGATGAGCAATTGGAATTCCTCTAAAAGTAATTGAAAATAGTTTA
ACTGCAATCTGATTAAGATAGAAATTCTAAACAGGCAATTCAAGAGAGGGATTAGGCCAT
AAGTTCATATAAATAAAGACTTTTTGATTTATAAGAAAATGAAAAGAAATTGCTCAGCGA
AAAGTCTTAGCTTCAGACTAGAATTGAGTTACTTGAAGAATAATAAAATTCTAACAAAGT
TGCAATCTTAGAAAAGCAAACCACTCTTCATTAAGGTTATAAACTCACATTTTTAAATAA
CGAATTAAAAGAATCTCTTATTGAACAACCTTCAAATGTATTTTAATAAGAAAACGGGGT
TAATAAAAATAAAATATATAGCTTGGATAATAATTAGATATAAAAGAATAATACTTCCCT
TTTAAGTAAATTTAAGTAAGACCAATCTGATTAAGAGGTGTAAAATAATACATAACTTGC
TCAAAAAAACTAATAGGATATTTATTATAGTAACAAAATAAATCAGATAAAACCTGAAAC
ATTTTCAAATCAAGATCCTTCAGTTTATTTACAAGAAAGCTATGTATAAAGTTCACTTAA
TGCAAATAATATTTTAGGTTCAAAAAAGATTACAGATGAAGAGATTTTTATGATGTAAAA
AAGAGAAGCTGAAATTAAAAAAAAGAAATAGGAAGATGAACATTTAAAGAATTAATAATA
CATGTCAAGTTTGAATTACAATAAAATAATTTCTGATAAAAATAAAAATTAGAATTTAAT
GAATAATAAAATTATTGAGTAATCTAAAGAGAGTTCTATGGGATCTTTATTCAATAAGAC
AGCTTTTGATCAAAGCTCAATTAATAATCAAAGTAATGCTAGTGTAAGAATGGAATTAGA
AGATTCTAGTGCTGGATAACAGCTTTAAAGATATAAAGTAAGCTTAAATGATAGAAAAAT
GGTAGCTGAAAATAATCTAAATGCAACAGATGATTTATTAAAAAATAAAATGGAGCAGGA
AGATAATGAAAAAGAGAGTTCTGACTCTTAGTATAACTGGAATTTAAAAATCGATTTGAA
AAACAATAACGAATAATAGCAGCAATTTCCATAGGAAAGCAATAACAATAGTGGAAACAA
AAAGAGATTCAAAAAGTAACAATTAAAAAAATCAGATTCTTTTGCCACAAATGCAAACTA
GAATTCTGAATTAGAAAACACCACAGCTAGTGTTATTAGTTTACAAAACTAACAATAAAA
TTAAAATTATGAAATTTAAGATCAATAAAGCAATTAAATGCTCTTAGGTCAAAAGAAAAA
TTTAAATATTAATAAAGGTAGAGTTGCTAATAATAAATTTATGAAAGACATAGAATTAGA
TGATGAAAGTGGGGACAAAGACTAACCCTAATAAAATGATATATTTGCTCAGTTTCAATA
TAAAAAAAGAGGTACTTCAAACTAAGAAGTTCAAGTAGCTGAAACTTTTAACATGTATAA
CAAATAAAATTAGAGACCATTAAATCCTCGTAAAAAAGAAGATAGAGAAAAATTAAAAAC
TTTTAATTGTGGCAGATGCAAAGCCGCATATCAAACTTTGGGAGAAGAATTCTGTGGTAA
TTGTTCTAAACACAGATCAGAATACACGCCTCCTCACACTCCCGAAGCCCATTATTAAAA
ATTTTCCTAGTCAGAAGATGATTCTAATTGAAAAAAAGAGTGCTAAAAACATATTTTAAA
ATTCAAATAAATGAATGAATAAATTAATTCTTAATGAGATTTCAAAATTGATAAATAAAA
TAATTAAATAATAAAATTAATAAAATAAATATGAAAATCACCAAAATTATTAATATAAAA
TGAATGTTCTTATTTAAATTAATTCTATTTTCTATTGTGTTTTTTATTTGTACTATCTTT
ATAGTTGTTATAATATATCTAAAAGAG
>TTHERM_00460480(protein)
MLGLEQNCFIKEYRKQVDKEALNQIQDAALSNLMNAFGQMNQTYSVLFKDICLTFGALLS
IKEELKDYRQEMFQKQSEKIIQEKIQKEMIQQKDEQLEFLQKQLKIVQLQSDQDRNSKQA
IQERDQAISSYKQRLFDLQENEKKLLSEKSQLQTRIELLEEQQNSNKVAILEKQTTLHQG
YKLTFLNNELKESLIEQPSNVFQQENGVNKNKIYSLDNNQIQKNNTSLLSKFKQDQSDQE
VQNNTQLAQKNQQDIYYSNKINQIKPETFSNQDPSVYLQESYVQSSLNANNILGSKKITD
EEIFMMQKREAEIKKKKQEDEHLKNQQYMSSLNYNKIISDKNKNQNLMNNKIIEQSKESS
MGSLFNKTAFDQSSINNQSNASVRMELEDSSAGQQLQRYKVSLNDRKMVAENNLNATDDL
LKNKMEQEDNEKESSDSQYNWNLKIDLKNNNEQQQQFPQESNNNSGNKKRFKKQQLKKSD
SFATNANQNSELENTTASVISLQNQQQNQNYEIQDQQSNQMLLGQKKNLNINKGRVANNK
FMKDIELDDESGDKDQPQQNDIFAQFQYKKRGTSNQEVQVAETFNMYNKQNQRPLNPRKK
EDREKLKTFNCGRCKAAYQTLGEEFCGNCSKHRSEYTPPHTPEAHYQKFSQSEDDSN
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
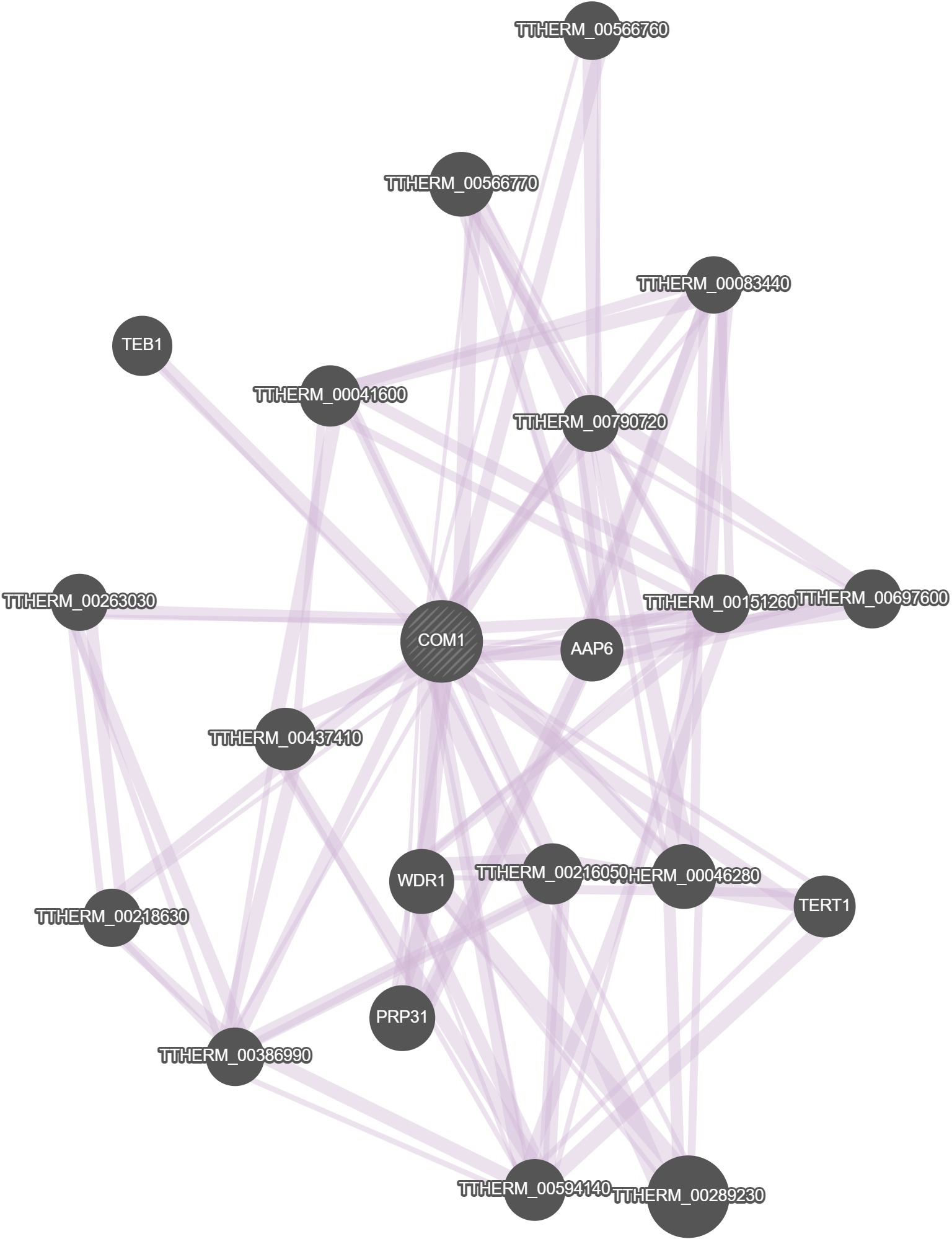
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences