Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00473180Standard Name
NRK37 (NIMA-Related Kinase )Aliases
PreTt02218 | 57.m00157 | 3709.m00018Description
NRK37 Serine/Threonine kinase domain protein; NIMA-Related Kinase; NEK Ser/Thr protein kinase family, NIMA subfamilyGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- aleurone grain membrane (IEA) | GO:0032578
- cortical granule (IEA) | GO:0060473
- CURI complex (IEA) | GO:0032545
- EGFR-Grb2-Sos complex (IEA) | GO:0070620
- gamma-catenin-TCF7L2 complex (IEA) | GO:0071665
- Golgi apparatus (IEA) | GO:0005794
- guanyl-nucleotide exchange factor complex (IEA) | GO:0032045
- multivesicular body (IEA) | GO:0005771
- obsolete chromosome segregation-directing complex (IEA) | GO:0043913
- obsolete farnesyl-diphosphate farnesyl transferase complex (IEA) | GO:0008051
- protein storage vacuole (IEA) | GO:0000326
- subrhabdomeral cisterna (IEA) | GO:0016029
- vacuolar HOPS complex (IEA) | GO:1902500
- vacuolar membrane (IEA) | GO:0005774
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 11beta-hydroxyprogesterone binding (IEA) | GO:1903879
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- ABC-type amino acid transporter activity (IEA) | GO:0015424
- ABC-type oligosaccharide transporter activity (IEA) | GO:0015422
- aromatase activity (IEA) | GO:0008402
- arsenate reductase (glutaredoxin) activity (IEA) | GO:0008794
- axon guidance receptor activity (IEA) | GO:0008046
- cAMP-dependent protein kinase regulator activity (IEA) | GO:0008603
- CCR10 chemokine receptor binding (IEA) | GO:0031735
- CCR11 chemokine receptor binding (IEA) | GO:0031736
- Cys-tRNA(Pro) hydrolase activity (IEA) | GO:0043907
- ferric enterobactin:proton symporter activity (IEA) | GO:0015345
- ferric triacetylfusarinine C:proton symporter activity (IEA) | GO:0015346
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- lysine:proton symporter activity (IEA) | GO:0015493
- metarhodopsin binding (IEA) | GO:0016030
- methane monooxygenase [NAD(P)H] activity (IEA) | GO:0015049
- microtubule severing ATPase activity (IEA) | GO:0008568
- myoinhibitory hormone activity (IEA) | GO:0016085
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete diazepam binding inhibitor activity (IEA) | GO:0008025
- obsolete phosphatidylinositol 3-kinase, class I, catalyst activity (IEA) | GO:0015072
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete type II protein secretor activity (IEA) | GO:0015447
- obsolete zinc, iron permease activity (IEA) | GO:0015342
- phosphatidylcholine:cardiolipin O-linoleoyltransferase activity (IEA) | GO:0032577
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- prenyltransferase activity (IEA) | GO:0004659
- protein geranylgeranyltransferase activity (IEA) | GO:0004661
- protein-glutamic acid ligase activity (IEA) | GO:0070739
- purinergic nucleotide receptor activity (IEA) | GO:0001614
- pyrimidine nucleotide-sugar transmembrane transporter activity (IEA) | GO:0015165
- siderophore uptake transmembrane transporter activity (IEA) | GO:0015344
- silver ion transmembrane transporter activity (IEA) | GO:0015080
- sodium-independent organic anion transmembrane transporter activity (IEA) | GO:0015347
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- sterol 14-demethylase activity (IEA) | GO:0008398
- thyroid hormone transmembrane transporter activity (IEA) | GO:0015349
Biological Process
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- 5alpha,9alpha,10beta-labda-8(20),13-dien-15-yl diphosphate biosynthetic process (IEA) | GO:1901949
- bronchiole development (IEA) | GO:0060435
- bronchus development (IEA) | GO:0060433
- bud dilation involved in lung branching (IEA) | GO:0060503
- carbon fixation (IEA) | GO:0015977
- cell migration in diencephalon (IEA) | GO:0061381
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to carbendazim (IEA) | GO:0072762
- cellular response to cocaine (IEA) | GO:0071314
- cellular response to nicotine (IEA) | GO:0071316
- coenzyme A biosynthetic process (IEA) | GO:0015937
- endoplasmic reticulum to cytosol transport (IEA) | GO:1903513
- energy derivation by oxidation of reduced inorganic compounds (IEA) | GO:0015975
- epithelium development (IEA) | GO:0060429
- fatty acid transmembrane transport (IEA) | GO:1902001
- fatty-acyl-CoA transport (IEA) | GO:0015916
- female courtship behavior (IEA) | GO:0008050
- flower phyllotactic patterning (IEA) | GO:0060773
- glycolytic process from glycerol (IEA) | GO:0061613
- hair cycle process (IEA) | GO:0022405
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- inferior colliculus development (IEA) | GO:0061379
- laminaritriose transport (IEA) | GO:2001097
- leaf phyllotactic patterning (IEA) | GO:0060772
- light absorption (IEA) | GO:0016037
- lung connective tissue development (IEA) | GO:0060427
- lung epithelium development (IEA) | GO:0060428
- lung pattern specification process (IEA) | GO:0060432
- lung saccule development (IEA) | GO:0060430
- maintenance of endoplasmic reticulum location involved in endoplasmic reticulum polarization at cell division site (IEA) | GO:0061167
- maltotetraose transport (IEA) | GO:2001099
- mesonephric juxtaglomerular apparatus development (IEA) | GO:0061212
- mesonephric macula densa development (IEA) | GO:0061220
- mesonephric nephron development (IEA) | GO:0061215
- mesonephric smooth muscle tissue development (IEA) | GO:0061214
- mesonephros development (IEA) | GO:0001823
- microtubule depolymerization (IEA) | GO:0007019
- mitochondrial DNA catabolic process (IEA) | GO:0032043
- mitochondrial membrane organization (IEA) | GO:0007006
- mitotic anaphase A (IEA) | GO:0000091
- mitotic DNA replication DNA ligation (IEA) | GO:1902973
- mitotic metaphase (IEA) | GO:0000089
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of bacterial-type flagellum-dependent cell motility (IEA) | GO:1902201
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of heart growth (IEA) | GO:0061117
- negative regulation of interleukin-4-mediated signaling pathway (IEA) | GO:1902215
- negative regulation of leukocyte cell-cell adhesion (IEA) | GO:1903038
- negative regulation of male mating behavior (IEA) | GO:1902436
- negative regulation of mesonephros development (IEA) | GO:0061218
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of protein processing (IEA) | GO:0010955
- negative regulation of telomere single strand break repair (IEA) | GO:1903824
- neutral lipid metabolic process (IEA) | GO:0006638
- nitrate transmembrane transport (IEA) | GO:0015706
- obsolete fertilization, exchange of chromosomal proteins (IEA) | GO:0035042
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete negative regulation of adhesion of symbiont to host epithelial cell (IEA) | GO:1905227
- obsolete negative regulation of response to extracellular stimulus (IEA) | GO:0032105
- obsolete positive regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process (IEA) | GO:1902964
- obsolete regulation of glucose-6-phosphatase activity (IEA) | GO:0032114
- obsolete response to lipoic acid (IEA) | GO:1990479
- ommochrome biosynthetic process (IEA) | GO:0006727
- peripheral tolerance induction (IEA) | GO:0002465
- peripheral tolerance induction to self antigen (IEA) | GO:0002466
- phagocytosis (IEA) | GO:0006909
- photoreceptor cell morphogenesis (IEA) | GO:0008594
- phyllotactic patterning (IEA) | GO:0060771
- pigment accumulation in tissues in response to UV light (IEA) | GO:0043479
- placenta blood vessel development (IEA) | GO:0060674
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of actin-dependent ATPase activity (IEA) | GO:1904623
- positive regulation of anti-Mullerian hormone signaling pathway (IEA) | GO:1902614
- positive regulation of double-strand break repair via homologous recombination (IEA) | GO:1905168
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of epithelial cell proliferation involved in lung bud dilation (IEA) | GO:0060504
- positive regulation of hair placode formation (IEA) | GO:0061169
- positive regulation of interleukin-4-mediated signaling pathway (IEA) | GO:1902216
- positive regulation of intestinal epithelial cell development (IEA) | GO:1905300
- positive regulation of male mating behavior (IEA) | GO:1902437
- positive regulation of peroxisome proliferator activated receptor signaling pathway (IEA) | GO:0035360
- positive regulation of protein processing (IEA) | GO:0010954
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of translational initiation in response to starvation (IEA) | GO:0071264
- positive regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903691
- post-chaperonin tubulin folding pathway (IEA) | GO:0007023
- premeiotic DNA replication DNA duplex unwinding (IEA) | GO:1902970
- priming of cellular response to stress (IEA) | GO:0080136
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein K27-linked deubiquitination (IEA) | GO:1990167
- protein localization to old growing cell tip (IEA) | GO:1903858
- pyrimidine deoxyribonucleotide biosynthetic process (IEA) | GO:0009221
- pyrimidine ribonucleotide catabolic process (IEA) | GO:0009222
- quercetin catabolic process (IEA) | GO:1901733
- quercetin metabolic process (IEA) | GO:1901732
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of L-arginine import into cell (IEA) | GO:1902826
- regulation of male mating behavior (IEA) | GO:1902435
- regulation of mesonephros development (IEA) | GO:0061217
- regulation of positive chemotaxis to cAMP (IEA) | GO:0061118
- regulation of protein localization to cell division site (IEA) | GO:1901900
- response to D-galactose (IEA) | GO:1905377
- response to indole-3-methanol (IEA) | GO:0071680
- response to leukotriene B4 (IEA) | GO:1905389
- response to methamphetamine hydrochloride (IEA) | GO:1904313
- response to transition metal nanoparticle (IEA) | GO:1990267
- ribosomal large subunit export from nucleus (IEA) | GO:0000055
- rRNA editing (IEA) | GO:0016548
- sperm aster formation (IEA) | GO:0035044
- sterol metabolic process (IEA) | GO:0016125
- superior colliculus development (IEA) | GO:0061380
- synaptic vesicle docking (IEA) | GO:0016081
- synaptic vesicle priming (IEA) | GO:0016082
- tetrasaccharide transport (IEA) | GO:2001098
- triterpenoid catabolic process (IEA) | GO:0016105
- visual behavior (IEA) | GO:0007632
- xanthophyll biosynthetic process (IEA) | GO:0016123
- xanthophyll catabolic process (IEA) | GO:0016124
 Domains
Domains
- ( PF00069 ) Protein kinase domain
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
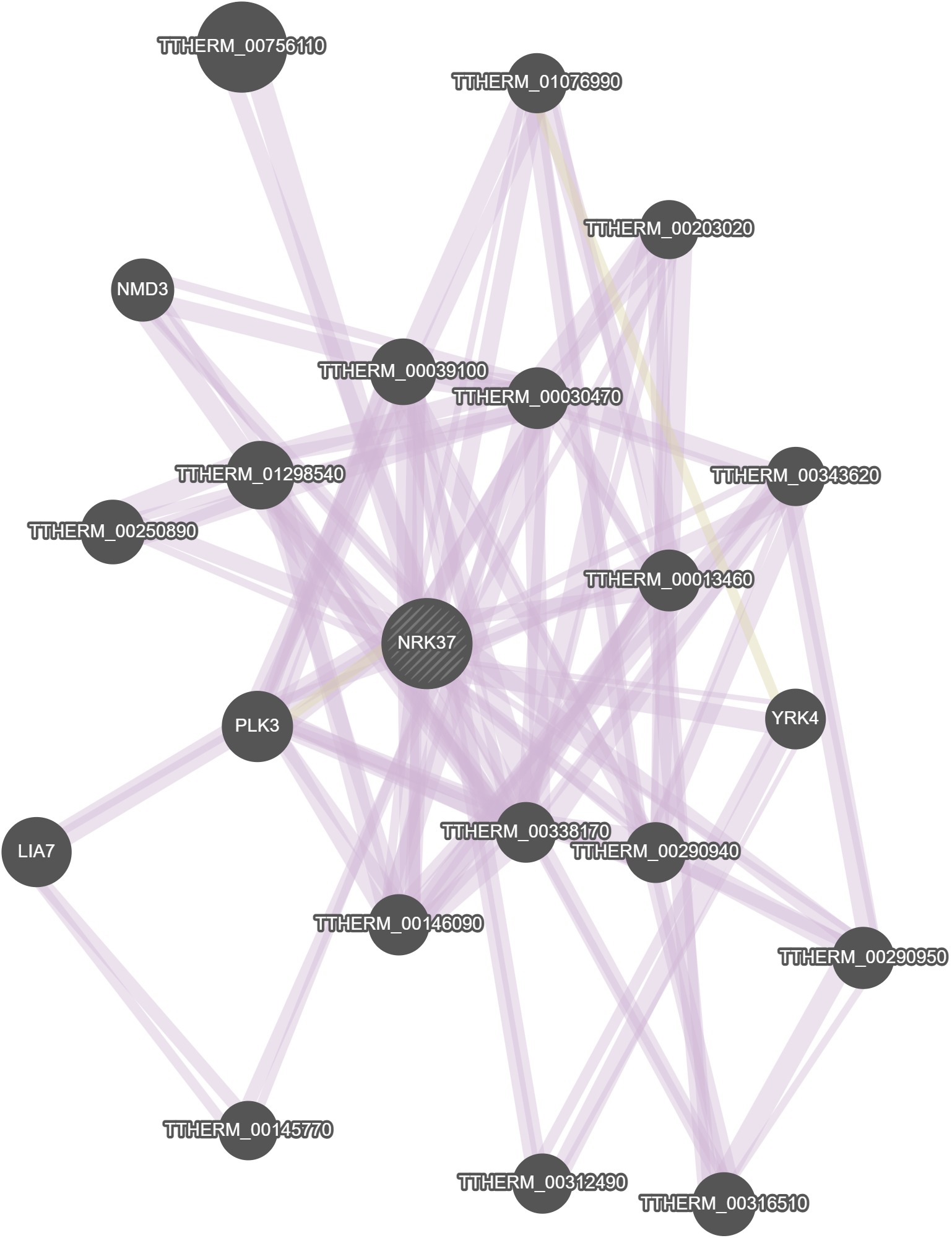
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:16611747: Wloga D, Camba A, Rogowski K, Manning G, Jerka-Dziadosz M, Gaertig J (2006) Members of the NIMA-related kinase family promote disassembly of cilia by multiple mechanisms. Molecular biology of the cell 17(6):2799-810
 Sequences
Sequences
>TTHERM_00473180(coding)
ATGGGATAGGAATTAAGAGATTTTGAAATTTTAGATAAATTAGGAGAAGGAGCATACAGT
TCTGTTTATAAAGTTAAGCGGTAGTCAGATGGTAAAATATATGCGCTAAAGAGAGTAATG
CTAGGTACATTAACTGAATAAGAAAAACAAAATGCATTAAATGAAGTCAGGATTATTGCT
TCTATCCGTCATCCAAATATAGTGAATTTCAAACAATGCTTTATAGACACACACTTAAAT
TGTTTATGTATTATCATGGAGTATGTAGATAACGGGGATTTACTATCTCATATTAACAAG
CACATTAAGAACGGTACTTACTTTTAAGAGGAAGAGATTTGGAATGTATTAATATAAGTT
GTAAAGGGATTACAGAGCCTCCATTAATTGAGCATTTATCATCGAGACTTGAAAAGTGCA
AATGTGTTTATGAATAAAGACGGTTCTATTAAGTTAGGTGATTTTAATGTTTCTAAAATA
TGTAAATAAGGTCTTTTGTACACTTAGACGGGTACGCCTTATTATGCAAGTCCTGAAGTT
TGGAGGGATTTACCATATGATAGCAAGGCAGATATATGGTCTGTTGGTTGCGTGATATAT
GAAATTTGTGCATTAAAACCCCCATTTAGAGCAGAAAATATGCCTTCTTTGTTTAAAAAA
GTAGTGAAAGGATAGTACCAAAAAATACCTTCTTTTTATTCTCAGTAGTTGAGCTAAATA
ATTAGATAAATGCTTTAAGTGTCTCCTCATTTACGTCCTAGCTGCAAAAAAATATTAGAA
GATGAAGTAGTATAAAAACATATGAATGATAAACATCTTGAAGAACCAGATGAACAAGAA
AGATCTATCCTTATAAATACAATAAAATTTCCTTAGGCTTTTAAAAATAAATATGCTAAA
ATAGAATTTCCAAAACCTAATTATCACCCTGTAAAAATAAGAAAAAGAATTTCAAGCAAC
ACCCGCTTGATAACAACTTTTGATTAAAGTGAATCCATTCTCTAATGCACTAACAAGAGC
ATGATAGAAGAATCATCTCATAAAATGCTCACAGAAAATTCTACTCATTAACCACCTTCA
AGCTAAAGACATCACTCTTAAAGATAAATATAAAACGAAGCTAGTATACTTTCAATGATA
GGTATTGAAAATACTAATATTAATTCTCATAGATAACAAAAGATAAATAAAAAAAATAGC
TATTGTTTATAACAATAAGATAAAATAGAATTATAAGAGGGAATTCAATAAAATAAAAAA
GATTATATAAATATTAACTAGCATGTAAATTATATCTAATAAATTGATAAAATAAGCAAC
ATAGAATTGAATGACTAGCTATTAGCCTCGTCTTAATCAAAAAAAAAAACAAATGCTTCT
CAATAGAATATTAATGGTTTAAATCAACAAAATATTATTCAAGGAAATTAACATAATGAA
GATTTATTTAGTGAATATTAGTCCAAAGCACTTATTATAGACAATTCTAAGAGTGTTTCT
CTTGCAGATGGTGAAAGAAAGATATAAAAATAGAGTATTTCAACAAATGAGAGCCGAAGA
AAATCTATTCATGCTAGATAATCAAGTAGTCTATCATCATAAGTTCAACCAAAAAATTAT
TCTTCAATAAATAAATAAAAATAGCTTTACAAAATATTAAAACAACCAATCAGCATAGTT
AAATAAATTCAACAATAACAATCACCATCTATAAATAATTAAAATGCTTAAACGAATTAG
AATATCATTAATTCATATAATAACAGTTTATTACTAAATTAATCTATAAACAATTAATAA
TATCCATAAAAAAATATTATCAAAAACCAAAGCATTTCATCTGTTTTAAGGGAAAGTTTG
ATTGCTCTAGAGAAAGATATAGCCAAAAGCACGAAAGGCTAAAGAGCCTCTATATAATAA
AACGATAGTAAAGAAATTAGCCATGCTAAAGGAGAATAAATTTAAAGAGATAATGAAAAG
TAAGTCTCAGTAAGTTATTCACGCTCTCACAAAAATTCTTATCACGTAGCACCTGATGGA
GTTCAAAAAAATGAAGAGGATTTTATAAATGGATAGTTGGACTCTATAGCATAATAAAAT
AAAATACAAAGTTATAAAGATAGAAATATTATACCAGATAAAGAAAATTTAAATGATATC
TAATATGTCTCCTAATTATAAACTACTTCTAAAACTACAAAAAACATGAAATAAAATTCT
AAATTAAATTATGAAAAAAGTAAAAGTAGAGAAATTAATCAAAATAGATAAAGCACACAT
TTGTAAGTAATATAAAATGGCTAAGTGAATTAGCACAGAATACCCACGCTATATGAATTA
GATAAATCTCCTGTAGCATATAATGAAGGAAGAAAAAAGAGTAAATCAATTTTCGAAAAA
AACAGTACAACAGTTACAGTCCCTGACTAATACATGAATTTAGAATATAAATATAAAAAA
GAGAAAATGTTAGAAAGCTCCTCAAGACAATAAATAGATAATAATGAAAACACTGAAAAT
ATATTAATAAAAAAACAAATTAAATATTCTGGTGAAACTCCTGAAAAGTTAATAATTCTA
CCAATAATTAACCCTGTTAAAAATACTAAATAAGATAGAAGTATTTCGGTTTAGAGAAAA
TACAGTCCAATATATTCACAAAATAATAACAGGGAAAATATTTTAAATAGGAATGGGCAC
ATATTTTAAGAAAACGATGTTAAAAAGAAGTTACTTTATAATGAAAATGAATAAAAAATA
TTAGATCCAAGTTAAATGCATTTAAATTAAAATAACTTATACATAAATAGTCACAAATAA
AATTACTACCACAATAAGGTCTCTTATTAGCAAACTCCTTTAGCATAAGATAAATATAAT
ATTTAATTAGATATGGAAACAATTGGTAATTATAAAAGAATCCATAGTAATTCAATCAAA
CCTGCACATAAAAATTCTCTATCTCCTTAGAATTACAGAAACAACCAGCAAAGATTAATT
TGA>TTHERM_00473180(gene)
TGAATCAATTAATACCAAAAAAATTCTAGATGGCACTAAAAAATTTATGATAAAAATAAA
AATCAGGGCTTAAAATTTATAGTTAGTATAGTATGAAAAATAATAAGATAAGTAGTTGTT
AAAAAAATAGCAAATGAATTTGTGAAGATAAGAAATCTATTGAAGTAACTAATAATAAGA
AAACAAAATATAATAACAATCACATTAAATCAATCAAATACTTTAAAAGTTTACTGCCAA
TAAAATTTGATTGATTGATTAATGGGATAGGAATTAAGAGATTTTGAAATTTTAGATAAA
TTAGGAGAAGGAGCATACAGTTCTGTTTATAAAGTTAAGCGGTAGTCAGATGGTAAAATA
TATGCGCTAAAGAGAGTAATGCTAGGTACATTAACTGAATAAGAAAAACAAAATGCATTA
AATGAAGTCAGGATTATTGCTTCTATCCGTCATCCAAATATAGTGAATTTCAAACAATGC
TTTATAGACACACACTTAAATTGTTTATGTATTATCATGGAGTATGTAGATAACGGGGAT
TTACTATCTCATATTAACAAGCACATTAAGAACGGTACTTACTTTTAAGAGGAAGAGATT
TGGAATGTATTAATATAAGTTGTAAAGGGATTACAGAGCCTCCATTAATTGAGCATTTAT
CATCGAGACTTGAAAAGTGCAAATGTGTTTATGAATAAAGACGGTTCTATTAAGTTAGGT
GATTTTAATGTTTCTAAAATATGTAAATAAGGTCTTTTGTACACTTAGACGGGTACGCCT
TATTATGCAAGTCCTGAAGTTTGGAGGGATTTACCATATGATAGCAAGGCAGATATATGG
TCTGTTGGTTGCGTGATATATGAAATTTGTGCATTAAAACCCCCATTTAGAGCAGAAAAT
ATGCCTTCTTTGTTTAAAAAAGTAGTGAAAGGATAGTACCAAAAAATACCTTCTTTTTAT
TCTCAGTAGTTGAGCTAAATAATTAGATAAATGCTTTAAGTGTCTCCTCATTTACGTCCT
AGCTGCAAAAAAATATTAGAAGATGAAGTAGTATAAAAACATATGAATGATAAACATCTT
GAAGAACCAGATGAACAAGAAAGATCTATCCTTATAAATACAATAAAATTTCCTTAGGCT
TTTAAAAATAAATATGCTAAAATAGAATTTCCAAAACCTAATTATCACCCTGTAAAAATA
AGAAAAAGAATTTCAAGCAACACCCGCTTGATAACAACTTTTGATTAAAGTGAATCCATT
CTCTAATGCACTAACAAGAGCATGATAGAAGAATCATCTCATAAAATGCTCACAGAAAAT
TCTACTCATTAACCACCTTCAAGCTAAAGACATCACTCTTAAAGATAAATATAAAACGAA
GCTAGTATACTTTCAATGATAGGTATTGAAAATACTAATATTAATTCTCATAGATAACAA
AAGATAAATAAAAAAAATAGCTATTGTTTATAACAATAAGATAAAATAGAATTATAAGAG
GGAATTCAATAAAATAAAAAAGATTATATAAATATTAACTAGCATGTAAATTATATCTAA
TAAATTGATAAAATAAGCAACATAGAATTGAATGACTAGCTATTAGCCTCGTCTTAATCA
AAAAAAAAAACAAATGCTTCTCAATAGAATATTAATGGTTTAAATCAACAAAATATTATT
CAAGGAAATTAACATAATGAAGATTTATTTAGTGAATATTAGTCCAAAGCACTTATTATA
GACAATTCTAAGAGTGTTTCTCTTGCAGATGGTGAAAGAAAGATATAAAAATAGAGTATT
TCAACAAATGAGAGCCGAAGAAAATCTATTCATGCTAGATAATCAAGTAGTCTATCATCA
TAAGTTCAACCAAAAAATTATTCTTCAATAAATAAATAAAAATAGCTTTACAAAATATTA
AAACAACCAATCAGCATAGTTAAATAAATTCAACAATAACAATCACCATCTATAAATAAT
TAAAATGCTTAAACGAATTAGAATATCATTAATTCATATAATAACAGTTTATTACTAAAT
TAATCTATAAACAATTAATAATATCCATAAAAAAATATTATCAAAAACCAAAGCATTTCA
TCTGTTTTAAGGGAAAGTTTGATTGCTCTAGAGAAAGATATAGCCAAAAGCACGAAAGGC
TAAAGAGCCTCTATATAATAAAACGATAGTAAAGAAATTAGCCATGCTAAAGGAGAATAA
ATTTAAAGAGATAATGAAAAGTAAGTCTCAGTAAGTTATTCACGCTCTCACAAAAATTCT
TATCACGTAGCACCTGATGGAGTTCAAAAAAATGAAGAGGATTTTATAAATGGATAGTTG
GACTCTATAGCATAATAAAATAAAATACAAAGTTATAAAGATAGAAATATTATACCAGAT
AAAGAAAATTTAAATGATATCTAATATGTCTCCTAATTATAAACTACTTCTAAAACTACA
AAAAACATGAAATAAAATTCTAAATTAAATTATGAAAAAAGTAAAAGTAGAGAAATTAAT
CAAAATAGATAAAGCACACATTTGTAAGTAATATAAAATGGCTAAGTGAATTAGCACAGA
ATACCCACGCTATATGAATTAGATAAATCTCCTGTAGCATATAATGAAGGAAGAAAAAAG
AGTAAATCAATTTTCGAAAAAAACAGTACAACAGTTACAGTCCCTGACTAATACATGAAT
TTAGAATATAAATATAAAAAAGAGAAAATGTTAGAAAGCTCCTCAAGACAATAAATAGAT
AATAATGAAAACACTGAAAATATATTAATAAAAAAACAAATTAAATATTCTGGTGAAACT
CCTGAAAAGTTAATAATTCTACCAATAATTAACCCTGTTAAAAATACTAAATAAGATAGA
AGTATTTCGGTTTAGAGAAAATACAGTCCAATATATTCACAAAATAATAACAGGGAAAAT
ATTTTAAATAGGAATGGGCACATATTTTAAGAAAACGATGTTAAAAAGAAGTTACTTTAT
AATGAAAATGAATAAAAAATATTAGATCCAAGTTAAATGCATTTAAATTAAAATAACTTA
TACATAAATAGTCACAAATAAAATTACTACCACAATAAGGTCTCTTATTAGCAAACTCCT
TTAGCATAAGATAAATATAATATTTAATTAGATATGGAAACAATTGGTAATTATAAAAGA
ATCCATAGTAATTCAATCAAACCTGCACATAAAAATTCTCTATCTCCTTAGAATTACAGA
AACAACCAGCAAAGATTAATTTGAAAGTTTCAAATAGCAATAAGTCACTCAGTATATTTT
TGATAAATTTTTCCAATCTTAAAGTGTAAATAGAAGTAATAAAAGGTTATAAAACTTATA
TTTAACTAATTAATTAATAAATTTATTTAGAGTTACATACATTTTTGAATAATTTTTTGA
TTGTTTGAAAATTTCAAATTTATATCTTAACAAAAAAAATCTAAAATAAAAGTGTATCAT
AATTAATAAGCTTAAAATAATTTACTCA>TTHERM_00473180(protein)
MGQELRDFEILDKLGEGAYSSVYKVKRQSDGKIYALKRVMLGTLTEQEKQNALNEVRIIA
SIRHPNIVNFKQCFIDTHLNCLCIIMEYVDNGDLLSHINKHIKNGTYFQEEEIWNVLIQV
VKGLQSLHQLSIYHRDLKSANVFMNKDGSIKLGDFNVSKICKQGLLYTQTGTPYYASPEV
WRDLPYDSKADIWSVGCVIYEICALKPPFRAENMPSLFKKVVKGQYQKIPSFYSQQLSQI
IRQMLQVSPHLRPSCKKILEDEVVQKHMNDKHLEEPDEQERSILINTIKFPQAFKNKYAK
IEFPKPNYHPVKIRKRISSNTRLITTFDQSESILQCTNKSMIEESSHKMLTENSTHQPPS
SQRHHSQRQIQNEASILSMIGIENTNINSHRQQKINKKNSYCLQQQDKIELQEGIQQNKK
DYININQHVNYIQQIDKISNIELNDQLLASSQSKKKTNASQQNINGLNQQNIIQGNQHNE
DLFSEYQSKALIIDNSKSVSLADGERKIQKQSISTNESRRKSIHARQSSSLSSQVQPKNY
SSINKQKQLYKILKQPISIVKQIQQQQSPSINNQNAQTNQNIINSYNNSLLLNQSINNQQ
YPQKNIIKNQSISSVLRESLIALEKDIAKSTKGQRASIQQNDSKEISHAKGEQIQRDNEK
QVSVSYSRSHKNSYHVAPDGVQKNEEDFINGQLDSIAQQNKIQSYKDRNIIPDKENLNDI
QYVSQLQTTSKTTKNMKQNSKLNYEKSKSREINQNRQSTHLQVIQNGQVNQHRIPTLYEL
DKSPVAYNEGRKKSKSIFEKNSTTVTVPDQYMNLEYKYKKEKMLESSSRQQIDNNENTEN
ILIKKQIKYSGETPEKLIILPIINPVKNTKQDRSISVQRKYSPIYSQNNNRENILNRNGH
IFQENDVKKKLLYNENEQKILDPSQMHLNQNNLYINSHKQNYYHNKVSYQQTPLAQDKYN
IQLDMETIGNYKRIHSNSIKPAHKNSLSPQNYRNNQQRLI