Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00475220Standard Name
RPB82 (RNA polymerases I, II,and III RNA common subunit RPB8B)Aliases
PreTt02321 | 57.m00260 | 3709.m00120 | RPB8BDescription
RPB82 RNA polymerase rpb8; RNA polymerase subunit ABC14.5, common to RNA polymerases I, II-and III; DNA-directed RNA polymerases I, II, and III subunit RPABC3Genome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- alpha5-beta1 integrin-osteopontin complex (IEA) | GO:0071128
- cellular bud neck septin structure (IEA) | GO:0000399
- CURI complex (IEA) | GO:0032545
- EGFR-Grb2-Sos complex (IEA) | GO:0070620
- growth cone membrane (IEA) | GO:0032584
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- prospore membrane spindle pole body attachment site (IEA) | GO:0070057
- SSL2-core TFIIH complex portion of holo TFIIH complex (IEA) | GO:0000443
- SSL2-core TFIIH complex portion of NEF3 complex (IEA) | GO:0000442
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-keto-3-deoxy-L-rhamnonate aldolase activity (IEA) | GO:0106099
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- all-trans retinol 3,4-desaturase activity (IEA) | GO:0061896
- arsenate reductase (glutaredoxin) activity (IEA) | GO:0008794
- carboxymethylenebutenolidase activity (IEA) | GO:0008806
- carboxyvinyl-carboxyphosphonate phosphorylmutase activity (IEA) | GO:0008807
- cardiolipin synthase activity (IEA) | GO:0008808
- cellulase activity (IEA) | GO:0008810
- cysteine-type deubiquitinase activity (IEA) | GO:0004843
- ferric triacetylfusarinine C:proton symporter activity (IEA) | GO:0015346
- melibiose binding (IEA) | GO:1903777
- MHC class II receptor activity (IEA) | GO:0032395
- microtubule severing ATPase activity (IEA) | GO:0008568
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- oligoxyloglucan reducing-end-specific cellobiohydrolase activity (IEA) | GO:0033945
- open form four-way junction DNA binding (IEA) | GO:0000401
- phosphatidylcholine:cardiolipin O-linoleoyltransferase activity (IEA) | GO:0032577
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase (ATP) activity (IEA) | GO:0004612
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- phosphopantothenate--cysteine ligase activity (IEA) | GO:0004632
- proteinase activated receptor binding (IEA) | GO:0031871
- pyrimidine nucleotide-sugar transmembrane transporter activity (IEA) | GO:0015165
- Ser(Gly)-tRNA(Ala) hydrolase activity (IEA) | GO:0043908
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- type 1 proteinase activated receptor binding (IEA) | GO:0031872
- type 3 proteinase activated receptor binding (IEA) | GO:0031874
Biological Process
- autophagy of mitochondrion (IEA) | GO:0000422
- basement membrane organization (IEA) | GO:0071711
- blood coagulation, intrinsic pathway (IEA) | GO:0007597
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to diacyl bacterial lipopeptide (IEA) | GO:0071726
- clathrin-independent pinocytosis (IEA) | GO:0006908
- coenzyme A biosynthetic process (IEA) | GO:0015937
- coenzyme A metabolic process (IEA) | GO:0015936
- effector-mediated activation of plant hypersensitive response by symbiont (IEA) | GO:0080185
- embryo implantation (IEA) | GO:0007566
- endoderm development (IEA) | GO:0007492
- epiboly involved in gastrulation with mouth forming second (IEA) | GO:0055113
- female pregnancy (IEA) | GO:0007565
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- internal genitalia morphogenesis (IEA) | GO:0035260
- learning (IEA) | GO:0007612
- learning or memory (IEA) | GO:0007611
- manganese ion transport (IEA) | GO:0006828
- mesodermal cell fate determination (IEA) | GO:0007500
- midgut development (IEA) | GO:0007494
- mitochondrial membrane organization (IEA) | GO:0007006
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of atrichoblast fate specification (IEA) | GO:0010060
- negative regulation of chemokine (C-C motif) ligand 20 production (IEA) | GO:1903885
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete age dependent increased protein content (IEA) | GO:0007573
- obsolete age-dependent yeast cell size increase (IEA) | GO:0007581
- obsolete cell aging (sensu Saccharomyces) (IEA) | GO:0007574
- obsolete extrachromosomal circular DNA accumulation involved in cell aging (IEA) | GO:0007580
- obsolete MAPK cascade involved in ascospore formation (IEA) | GO:1903695
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete negative regulation of RNA binding transcription factor activity (IEA) | GO:1905256
- obsolete negative regulation of SMAD protein complex assembly (IEA) | GO:0010991
- obsolete nucleolar fragmentation (IEA) | GO:0007576
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete senescence factor accumulation (IEA) | GO:0007579
- obsolete sodium/potassium transport (IEA) | GO:0006815
- ommochrome biosynthetic process (IEA) | GO:0006727
- parturition (IEA) | GO:0007567
- phagocytosis (IEA) | GO:0006909
- photoreceptor cell morphogenesis (IEA) | GO:0008594
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of bacterial-type flagellum assembly (IEA) | GO:1902210
- positive regulation of chemokine (C-C motif) ligand 20 production (IEA) | GO:1903886
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of response to water deprivation (IEA) | GO:1902584
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- prothoracic disc morphogenesis (IEA) | GO:0007470
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of bacterial-type flagellum assembly (IEA) | GO:1902208
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of chitin-based cuticle tanning (IEA) | GO:0007564
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of low-density lipoprotein particle clearance (IEA) | GO:0010988
- regulation of phosphorus utilization (IEA) | GO:0006795
- response to nutrient (IEA) | GO:0007584
- response to UV-B (IEA) | GO:0010224
- short-term memory (IEA) | GO:0007614
- smooth muscle adaptation (IEA) | GO:0014805
- snoRNA guided rRNA 2'-O-methylation (IEA) | GO:0000452
- sodium-independent prostaglandin transport (IEA) | GO:0071720
- sperm aster formation (IEA) | GO:0035044
- synaptic vesicle docking (IEA) | GO:0016081
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- triterpenoid catabolic process (IEA) | GO:0016105
- vesicle coating (IEA) | GO:0006901
- zinc ion transport (IEA) | GO:0006829
 Domains
Domains
- ( PF03870 ) RNA polymerase Rpb8
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
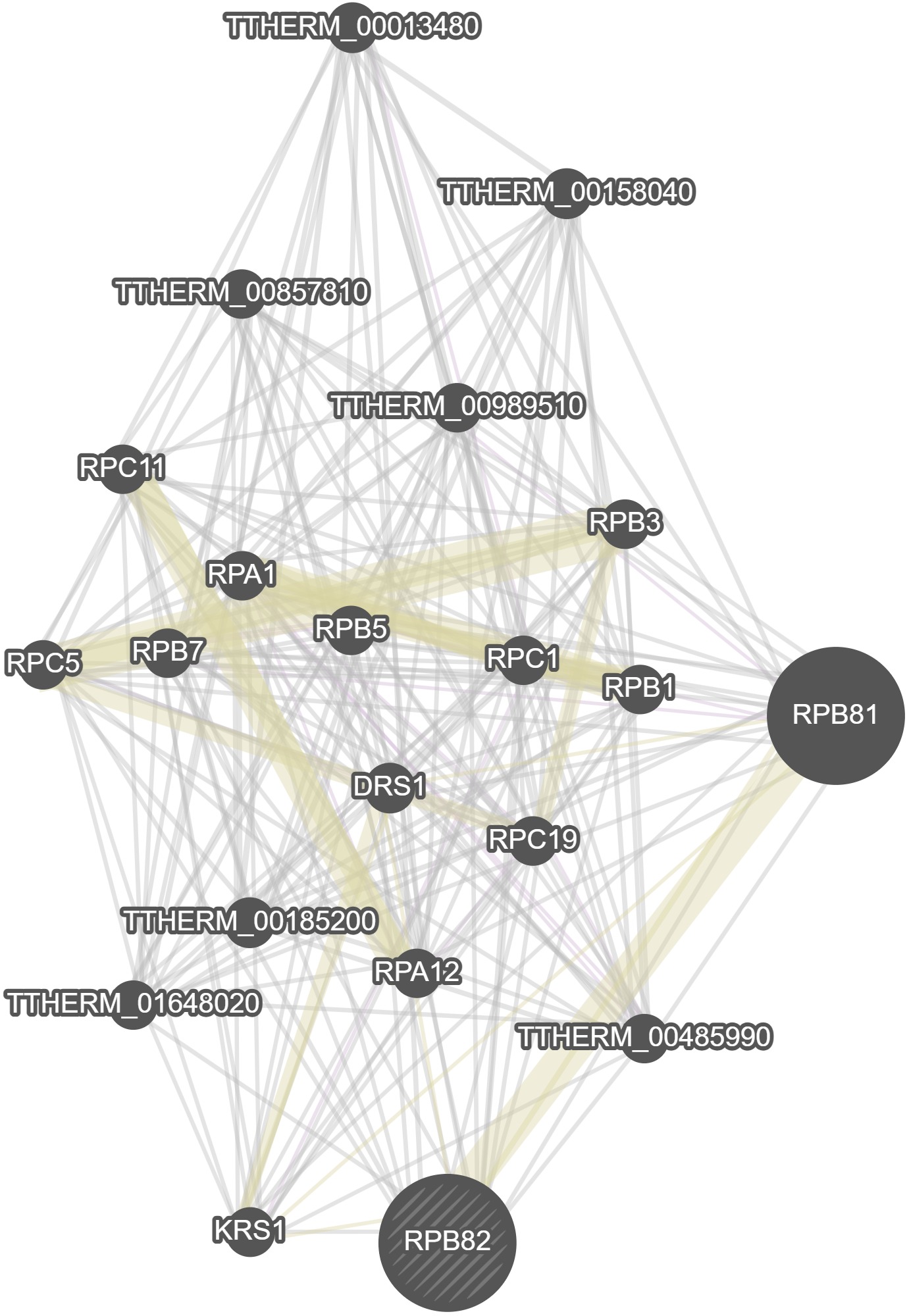
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00475220(coding)
ATGAATAATATTACACATATGACAGATTTTACTAGCGATGCTATTTTAGTTGATGATACT
TTCATAGCATCAAATATAATAAAAGAACCTAAATATGTTAGAACTTGGAGAGTAATTTTG
AATAATGAAACATTATCAATCGAGATAGATATCAATTTTGATCTCTTCCCTATGGAACAA
GACAAAAAATATTAGGTTATTCTCACTCAATCATTTTTACCAGAGTATAAAAAGAGCTTT
TCTTATGCCATGTATGGAAAGATATTTAAAATTGATTTAGAATAAGATTAATATTAAAGC
ATATATATTTCTTATGGAGGTCTTGTTATGAAAATGAGAGGTAAATTAAATGATATGAGC
TAAAGCATAAAAAATGAAACTCGTTGCTATATTTTACTAAGATAAATTTGA>TTHERM_00475220(gene)
AAATTTTTTATATACTTAAAGAATTACTAAAGTTTTTAATATTTACTGGAAGTTAAAGCT
AAAAATTAATTTGAAATGAATAATATTACACATATGACAGATTTTACTAGCGATGCTATT
TTAGTTGATGATACTTTCATAGCATCAAATATAATAAAAGAACCTAAATATGTTAGAACT
TGGAGAGTAATTTTGAATAATGAAACATTATCAATCGAGATAGATATCAATTTTGATCTC
TTCCCTATGGAACAAGACAAAAAATATTAGGTTATTCTCACTCAATCATTTTTACCAGAG
TATAAAAAGAGCTTTTCTTATGCCATGTATGGAAAGATATTTAAAATTGATTTAGAATAA
GATTAATATTAAAGCATATATATTTCTTATGGAGGTCTTGTTATGAAAATGAGAGGTAAA
TTAAATGATATGAGCTAAAGCATAAAAAATGAAACTCGTTGCTATATTTTACTAAGATAA
ATTTGAATAAAAAACTTTCTTAATTGATTTGGAAGTGCTTTTAAAATTATATATAAACTT
TTTTTATGTGTACTATGTAAATATTCTATCACAACTGATTAAAAAATTATAAATTAAATT
GATTTTAGTCTTTATCTAAAATTATCATTTTGCTTTTATTTTATAATAAGCTTAAAGCTC
TTAAAAA>TTHERM_00475220(protein)
MNNITHMTDFTSDAILVDDTFIASNIIKEPKYVRTWRVILNNETLSIEIDINFDLFPMEQ
DKKYQVILTQSFLPEYKKSFSYAMYGKIFKIDLEQDQYQSIYISYGGLVMKMRGKLNDMS
QSIKNETRCYILLRQI