Identifiers and Description
Identifiers and Description
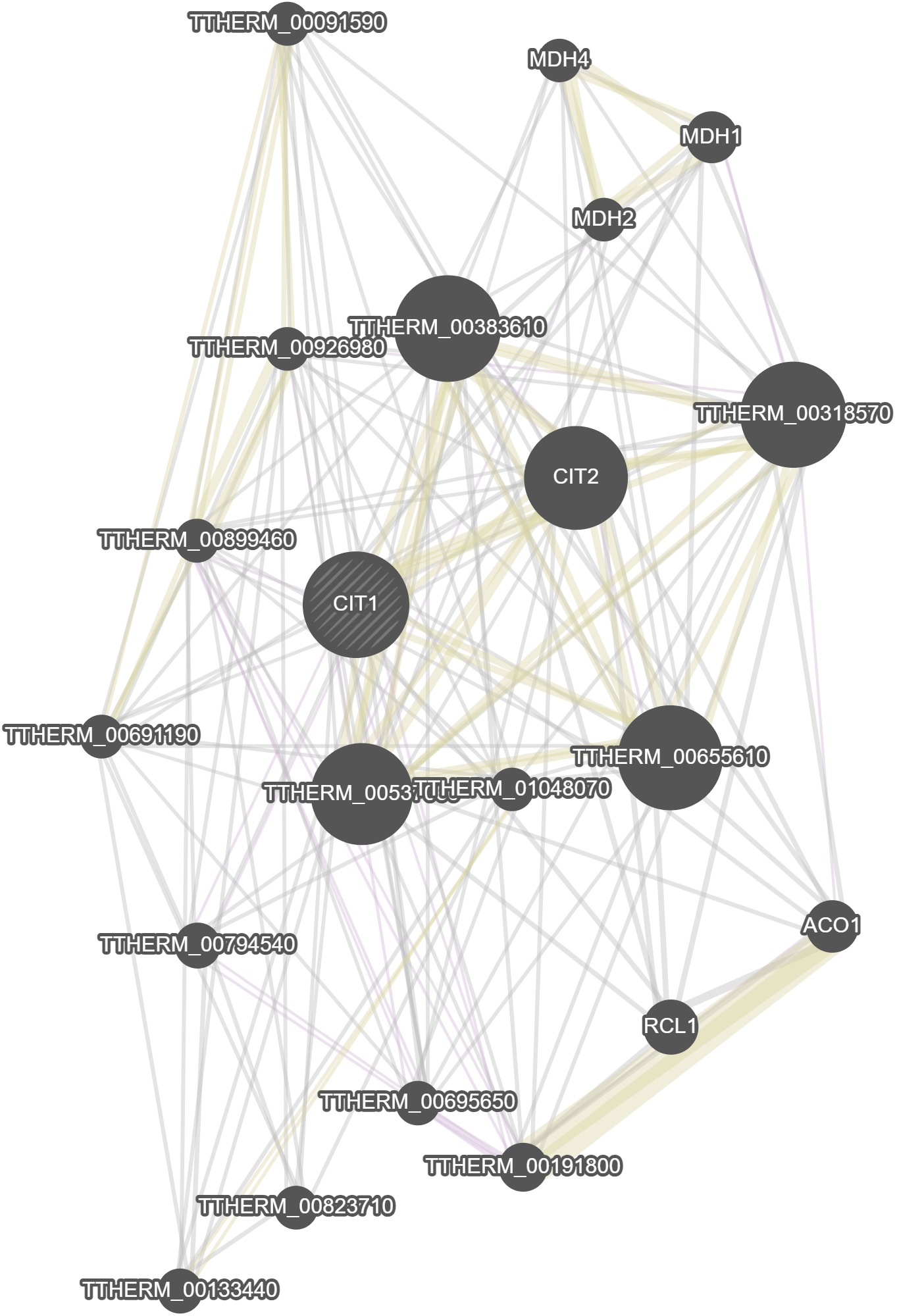
Gene Model Identifier
TTHERM_00529790Standard Name
CIT1 (CITrate synthase 1)Aliases
PreTt04648 | 67.m00163 | 3684.m00033Description
CIT1 citrate synthase I; Citrate synthase; bifunctional 14-nm filament-forming protein; structural protein involved in oral morphogenesis and pronuclear behavior during conjugation; citrate synthase activity decreased by polymerization and dephosphorylation; Citrate synthaseGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- mitochondrion (IDA) | GO:0005739
- oral apparatus (IDA) | GO:0031912
Molecular Function
- citrate (Si)-synthase activity (IDA) | GO:0004108
- heat shock protein binding (IPI) | GO:0031072
- structural constituent of cytoskeleton (IDA) | GO:0005200
 Domains
Domains
- ( PF00285 ) Citrate synthase, C-terminal domain
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:15170257: Mukai A, Endoh H (2004) Presence of a bacterial-like citrate synthase gene in Tetrahymena thermophila: recent lateral gene transfers (LGT) or multiple gene losses subsequent to a single ancient LGT? Journal of molecular evolution 58(5):540-9
- Ref:12025402: Kojima H, Numata O (2002) Enzymatic form and cytoskeletal form of bifunctional Tetrahymena 49kDa protein is regulated by phosphorylation. Zoological science 19(1):37-42
- Ref:11260259: Takeda T, Yoshihama I, Numata O (2001) Identification of Tetrahymena hsp60 as a 14-nm filament protein/citrate synthase-binding protein and its possible involvement in the oral apparatus formation. Genes to cells : devoted to molecular & cellular mechanisms 6(2):139-49
- Ref:9443816: Kojima H, Watanabe Y, Numata O (1997) The dual functions of Tetrahymena citrate synthase are due to the polymorphism of its isoforms. Journal of biochemistry 122(5):998-1003
- Ref:9268687: Takeda T, Watanabe Y, Numata O (1997) Direct demonstration of the bifunctional property of Tetrahymena 14-nm filament protein/citrate synthase following expression of the gene in Escherichia coli. Biochemical and biophysical research communications 237(2):205-10
- Ref:8575889: Numata O (1996) Multifunctional proteins in Tetrahymena: 14-nm filament protein/citrate synthase and translation elongation factor-1 alpha. International review of cytology 164( ):1-35
- Ref:7737352: Numata O, Yasuda T, Watanabe Y (1995) Tetrahymena intramitochondrial filamentous inclusions contain 14-nm filament protein/citrate synthase. Experimental cell research 218(1):123-31
- Ref:7592552: Takeda T, Kurasawa Y, Watanabe Y, Numata O (1995) Polymerization of highly purified Tetrahymena 14-nm filament protein/citrate synthase into filaments and its possible role in regulation of enzymatic activity. Journal of biochemistry 117(4):869-74
- Ref:8537311: Kojima H, Chiba J, Watanabe Y, Numata O (1995) Citrate synthase purified from Tetrahymena mitochondria is identical with Tetrahymena 14-nm filament protein. Journal of biochemistry 118(1):189-95
- Ref:1385189: Kurasawa Y, Numata O, Katoh M, Hirano H, Chiba J, Watanabe Y (1992) Identification of Tetrahymena 14-nm filament-associated protein as elongation factor 1 alpha. Experimental cell research 203(1):251-8
- Ref:1993043: Numata O, Takemasa T, Takagi I, Hirono M, Hirano H, Chiba J, Watanabe Y (1991) Tetrahymena 14-nm filament-forming protein has citrate synthase activity. Biochemical and biophysical research communications 174(2):1028-34
- Ref:3883198: Numata O, Sugai T, Watanabe Y (1985) Control of germ cell nuclear behaviour at fertilization by Tetrahymena intermediate filament protein. Nature 314(6007):192-4
- Ref:6401983: Fabregat I, Satrústegui J, Machado A (1983) Citrate synthase of Tetrahymena pyriformis: evolutionary and regulatory aspects. Archives of biochemistry and biophysics 220(2):354-60
- Ref:6414830: Numata O, Hirono M, Watanabe Y (1983) Involvement of Tetrahymena intermediate filament protein, a 49K protein, in the oral morphogenesis. Experimental cell research 148(1):207-20
- Ref:6807972: Numata O, Watanabe Y (1982) In vitro assembly and disassembly of 14-nm filament from Tetrahymena pyriformis. The protein component of 14-nm filament is a 49,000-Dalton protein. Journal of biochemistry 91(5):1563-73
- Ref:14168: Levy MR, McConkey CL (1977) Enzyme inactivation by a cellular neutral protease: enzyme specificity, effects of ligands on inactivation, and implications for the regulation of enzyme degradation. Journal of cellular physiology 90(2):253-63
 Sequences
Sequences
>TTHERM_00529790(coding)
ATGAGATCTATCAACTAACTTCTTAAGCAAGCTAGCTTATCCCAAAAGAGCCAATACAAC
TTTAGTCAAACCAACTTAAAGAAGGTTATTGCTGAAATTATTCCTCAAAAGCAAGCTGAA
TTAAAGGAAGTCAAGGAAAAGTATGGTGATAAGGTTGTTGGTTAATACACTGTCAAGTAA
GTCATCGGTGGTATGAGAGGTATGAAGGGTCTTATGAGTGATCTCTCTCGTTGCGACCCC
TATCAAGGTATTATTTTCAGAGGCTACACTATTCCTCAACTTAAGGAATTCTTGCCTAAG
GCTGATCCTAAGGCTGCTGACTAAGCCAACTAAGAACCTTTACCTGAAGGTATTTTCTGG
TTATTAATGACCGGTTAATTACCCACCCACGCTTAAGTCGATGCTCTTAAGCACGAATGG
TAAAACAGAGGTACCGTCAACTAAGACTGTGTCAACTTCATTTTGAACTTACCCAAGGAC
CTCCACTCCATGACCATGCTCTCTATGGCCCTCCTTTACCTCCAAAAGGATTCTAAGTTC
GCTAAGCTTTACGACGAAGGTAAGATCTCCAAGAAAGATTACTGGGAGCCCTTCTACGAA
GATTCCATGGACCTTATTGCTAAGATCCCCAGAGTTGCTGCTATTATCTACAGACATAAA
TACAGAGATTCCAAGTTAATTGACTCTGATTCTAAGTTAGATTGGGCTGGTAACTACGCT
CATATGATGGGTTTCGAATAACACGTCGTTAAGGAATGTATTAGAGGTTATTTGAGCATC
CACTGTGATCACGAAGGTGGTAATGTTTCTGCCCATACTACTCACTTGGTCGGTTCTGCT
CTTTCTGATCCTTATCTTTCTTACTCTGCTGGTGTTAACGGTCTTGCCGGTCCCCTCCAC
GGTCTTGCTAACTAAGAAGTCCTTAAGTGGTTACTCTAATTCATTGAAGAAAAGGGTACC
AAGGTTTCTGACAAGGACATTGAAGACTACGTTGATCACGTTATTTCTTCTGGAAGAGTC
GTTCCTGGTTACGGACACGCTGTCTTGAGAGATACCGATCCTAGATTCCACCATTAAGTC
GATTTCTCCAAGTTCCACTTGAAGGATGACTAAATGATCAAATTATTACATTAATGTGCT
GATGTTATCCCCAAGAAATTATTAACTTACAAGAAGATTGCTAATCCTTATCCCAATGTT
GATTGCCACTCTGGTGTTTTATTATACTCTTTGGGCTTAACTGAATATCAATATTACACT
GTTGTCTTCGCCGTTTCTAGAGCTCTTGGTTGTATGGCTAACCTCATCTGGTCTAGAGCT
TTCGGTTTACCCATTGAAAGACCTGGCTCTGCTGATCTTAAGTGGTTCCACGACAAGTAC
AGAGAATGA>TTHERM_00529790(gene)
AAAAAATAAGTAAAATAATCTTATTAAAAATATATCATCAAAAAATGAGATCTATCAACT
AACTTCTTAAGCAAGCTAGCTTATCCCAAAAGAGCCAATACAACTTTAGTCAAACCAACT
TAAAGAAGGTTATTGCTGAAATTATTCCTCAAAAGCAAGCTGAATTAAAGGAAGTCAAGG
AAAAGTATGGTGATAAGGTTGTTGGTTAATACACTGTCAAGTAAGTCATCGGTGGTATGA
GAGGTATGAAGGGTCTTATGAGTGATCTCTCTCGTTGCGACCCCTATCAAGGTATTATTT
TCAGAGGCTACACTATTCCTCAACTTAAGGAATTCTTGCCTAAGGCTGATCCTAAGGCTG
CTGACTAAGCCAACTAAGAACCTTTACCTGAAGGTATTTTCTGGTTATTAATGACCGGTT
AATTACCCACCCACGCTTAAGTCGATGCTCTTAAGCACGAATGGTAAAACAGAGGTACCG
TCAACTAAGACTGTGTCAACTTCATTTTGAACTTACCCAAGGACCTCCACTCCATGACCA
TGCTCTCTATGGCCCTCCTTTACCTCCAAAAGGATTCTAAGTTCGCTAAGCTTTACGACG
AAGGTAAGATCTCCAAGAAAGATTACTGGGAGCCCTTCTACGAAGATTCCATGGACCTTA
TTGCTAAGATCCCCAGAGTTGCTGCTATTATCTACAGACATAAATACAGAGATTCCAAGT
TAATTGACTCTGATTCTAAGTTAGATTGGGCTGGTAACTACGCTCATATGATGGGTTTCG
AATAACACGTCGTTAAGGAATGTATTAGAGGTTATTTGAGCATCCACTGTGATCACGAAG
GTGGTAATGTTTCTGCCCATACTACTCACTTGGTCGGTTCTGCTCTTTCTGATCCTTATC
TTTCTTACTCTGCTGGTGTTAACGGTCTTGCCGGTCCCCTCCACGGTCTTGCTAACTAAG
AAGTCCTTAAGTGGTTACTCTAATTCATTGAAGAAAAGGGTACCAAGGTTTCTGACAAGG
ACATTGAAGACTACGTTGATCACGTTATTTCTTCTGGAAGAGTCGTTCCTGGTTACGGAC
ACGCTGTCTTGAGAGATACCGATCCTAGATTCCACCATTAAGTCGATTTCTCCAAGTTCC
ACTTGAAGGATGACTAAATGATCAAATTATTACATTAATGTGCTGATGTTATCCCCAAGA
AATTATTAACTTACAAGAAGATTGCTAATCCTTATCCCAATGTTGATTGCCACTCTGGTG
TTTTATTATACTCTTTGGGCTTAACTGAATATCAATATTACACTGTTGTCTTCGCCGTTT
CTAGAGCTCTTGGTTGTATGGCTAACCTCATCTGGTCTAGAGCTTTCGGTTTACCCATTG
AAAGACCTGGCTCTGCTGATCTTAAGTGGTTCCACGACAAGTACAGAGAATGAACAAATT
GAACTATAAATTATTACTACTCTAATATTATTTTCGTCGATATAAATTTCTAATTAGTCT
TTAATAAAAGTTGTATCAATTCAACTAATATCAAAGCTTTATAATATTTTGTGTCAGTAT
CAAATGAGAAATATTAAAAGTTTTGACGATTTAATAAATAATTAAT>TTHERM_00529790(protein)
MRSINQLLKQASLSQKSQYNFSQTNLKKVIAEIIPQKQAELKEVKEKYGDKVVGQYTVKQ
VIGGMRGMKGLMSDLSRCDPYQGIIFRGYTIPQLKEFLPKADPKAADQANQEPLPEGIFW
LLMTGQLPTHAQVDALKHEWQNRGTVNQDCVNFILNLPKDLHSMTMLSMALLYLQKDSKF
AKLYDEGKISKKDYWEPFYEDSMDLIAKIPRVAAIIYRHKYRDSKLIDSDSKLDWAGNYA
HMMGFEQHVVKECIRGYLSIHCDHEGGNVSAHTTHLVGSALSDPYLSYSAGVNGLAGPLH
GLANQEVLKWLLQFIEEKGTKVSDKDIEDYVDHVISSGRVVPGYGHAVLRDTDPRFHHQV
DFSKFHLKDDQMIKLLHQCADVIPKKLLTYKKIANPYPNVDCHSGVLLYSLGLTEYQYYT
VVFAVSRALGCMANLIWSRAFGLPIERPGSADLKWFHDKYRE