Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00558460Standard Name
EBP1 (microtubule-Binding Protein 1)Aliases
PreTt07942 | 73.m00186 | 3735.m00080Description
EBP1 microtubule associated protein EB1 putative; Microtubule-binding protein, putative; Microtubule-associated protein RP/EBGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- chloroplast ribosome (IEA) | GO:0043253
- ciliary neurotrophic factor receptor complex (IEA) | GO:0070110
- Dbf4-dependent protein kinase complex (IEA) | GO:0031431
- endobrevin-SNAP-25-syntaxin-1a complex (IEA) | GO:0070048
- obsolete ectoplasm (IEA) | GO:0043265
- organellar chromatophore (IEA) | GO:0070111
- organellar chromatophore membrane (IEA) | GO:0070112
- subrhabdomeral cisterna (IEA) | GO:0016029
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-octaprenyl-6-methoxyphenol hydroxylase activity (IEA) | GO:0008681
- 2-oxoglutarate decarboxylase activity (IEA) | GO:0008683
- 2-oxopent-4-enoate hydratase activity (IEA) | GO:0008684
- 3-hydroxybutyryl-CoA dehydrogenase activity (IEA) | GO:0008691
- 3-octaprenyl-4-hydroxybenzoate carboxy-lyase activity (IEA) | GO:0008694
- 4-amino-4-deoxychorismate lyase activity (IEA) | GO:0008696
- A3 adenosine receptor activity, G-protein coupled (IEA) | GO:0001613
- ADP phosphatase activity (IEA) | GO:0043262
- antheraxanthin epoxidase activity (IEA) | GO:0052663
- bacterial-type RNA polymerase regulatory region DNA binding (IEA) | GO:0001017
- C-5 sterol desaturase activity (IEA) | GO:0000248
- carbamate kinase activity (IEA) | GO:0008804
- carotene beta-ring hydroxylase activity (IEA) | GO:0010291
- CDP-diacylglycerol diphosphatase activity (IEA) | GO:0008715
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) inhibitor activity (IEA) | GO:0008657
- ferric triacetylfusarinine C:proton symporter activity (IEA) | GO:0015346
- G protein-coupled neurotensin receptor activity (IEA) | GO:0016492
- inositol bisdiphosphate tetrakisphosphate diphosphatase activity (IEA) | GO:0052841
- interleukin-13 receptor activity (IEA) | GO:0016515
- L-aspartate oxidase activity (IEA) | GO:0008734
- L-carnitine CoA-transferase activity (IEA) | GO:0008735
- L(+)-tartrate dehydratase activity (IEA) | GO:0008730
- long-chain-3-hydroxyacyl-CoA dehydrogenase activity (IEA) | GO:0016509
- metarhodopsin binding (IEA) | GO:0016030
- mitochondrial promoter sequence-specific DNA binding (IEA) | GO:0001018
- obsolete actin polymerizing activity (IEA) | GO:0008651
- obsolete DNA topoisomerase IV activity (IEA) | GO:0008724
- obsolete isoleucine/valine:sodium symporter activity (IEA) | GO:0000258
- obsolete protein tagging activity (IEA) | GO:0008638
- obsolete RNA polymerase I transcription regulator recruiting activity (IEA) | GO:0001186
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete tachykinin (IEA) | GO:0008648
- obsolete tocotrienol omega-hydroxylase activity (IEA) | GO:0052872
- obsolete zinc, iron permease activity (IEA) | GO:0015342
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- plastid single-subunit type RNA polymerase binding (IEA) | GO:0001051
- preribosome binding (IEA) | GO:1990275
- rhamnulose-1-phosphate aldolase activity (IEA) | GO:0008994
- RNA polymerase III transcription regulatory region sequence-specific DNA binding (IEA) | GO:0001016
- rRNA (uridine-2'-O-)-methyltransferase activity (IEA) | GO:0008650
- rRNA methyltransferase activity (IEA) | GO:0008649
- salicylic acid glucosyltransferase (glucoside-forming) activity (IEA) | GO:0052640
- siderophore uptake transmembrane transporter activity (IEA) | GO:0015344
- single-subunit type RNA polymerase binding (IEA) | GO:0001050
- Swi5-Sfr1 complex binding (IEA) | GO:1905334
- TFIID-class transcription factor complex binding (IEA) | GO:0001094
- titin binding (IEA) | GO:0031432
- tubulin-glutamic acid ligase activity (IEA) | GO:0070740
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
- UDP-galactopyranose mutase activity (IEA) | GO:0008767
Biological Process
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- absorption of visible light (IEA) | GO:0016038
- analia development (IEA) | GO:0007487
- anatomical structure maturation (IEA) | GO:0071695
- branch elongation involved in ureteric bud branching (IEA) | GO:0060681
- bud dilation involved in lung branching (IEA) | GO:0060503
- calcium ion import into cytosol (IEA) | GO:1902656
- camera-type eye photoreceptor cell fate commitment (IEA) | GO:0060220
- carotenoid metabolic process (IEA) | GO:0016116
- cell migration in diencephalon (IEA) | GO:0061381
- cell-cell adhesion involved in sealing an epithelial fold (IEA) | GO:0060607
- cellular response to D-galactose (IEA) | GO:1905378
- cellular response to dopamine (IEA) | GO:1903351
- cellular response to nitrogen starvation (IEA) | GO:0006995
- cellular response to phosphate starvation (IEA) | GO:0016036
- ceramide phosphoethanolamine metabolic process (IEA) | GO:1905371
- coenzyme A biosynthetic process (IEA) | GO:0015937
- commissural neuron axon guidance (IEA) | GO:0071679
- definitive hemopoiesis (IEA) | GO:0060216
- detection of cell density by secreted molecule (IEA) | GO:0060247
- embryonic brain development (IEA) | GO:1990403
- endocardium formation (IEA) | GO:0060214
- epithelial cell proliferation involved in lung bud dilation (IEA) | GO:0060505
- epithelium development (IEA) | GO:0060429
- establishment of protein localization to prospore membrane (IEA) | GO:1902658
- establishment of spindle pole body localization (IEA) | GO:0070632
- ethylene biosynthetic process (IEA) | GO:0009693
- extrinsic apoptotic signaling pathway via death domain receptors (IEA) | GO:0008625
- fibroblast growth factor receptor signaling pathway (IEA) | GO:0008543
- germinal center formation (IEA) | GO:0002467
- heart trabecula morphogenesis (IEA) | GO:0061384
- hemangioblast cell differentiation (IEA) | GO:0060217
- hematopoietic stem cell differentiation (IEA) | GO:0060218
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- induction by virus of host reticulophagy (IEA) | GO:0140883
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- inferior colliculus development (IEA) | GO:0061379
- interleukin-12-mediated signaling pathway (IEA) | GO:0035722
- interleukin-15-mediated signaling pathway (IEA) | GO:0035723
- intraciliary retrograde transport (IEA) | GO:0035721
- iodide transmembrane transport (IEA) | GO:1904200
- L-methionine import into cell (IEA) | GO:1903813
- leucine import (IEA) | GO:0060356
- low-density lipoprotein particle mediated signaling (IEA) | GO:0055096
- lung connective tissue development (IEA) | GO:0060427
- lung pattern specification process (IEA) | GO:0060432
- lung saccule development (IEA) | GO:0060430
- Malpighian tubule tip cell differentiation (IEA) | GO:0061382
- mitotic cytokinetic cell separation (IEA) | GO:1902409
- mitotic metaphase (IEA) | GO:0000089
- mitotic S phase (IEA) | GO:0000084
- mitotic telophase (IEA) | GO:0000093
- mucopolysaccharide metabolic process (IEA) | GO:1903510
- N-alpha,N-alpha,N-alpha-trimethyl-L-histidine catabolic process (IEA) | GO:0052710
- N-alpha,N-alpha,N-alpha-trimethyl-L-histidine metabolic process (IEA) | GO:0052708
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of axon guidance (IEA) | GO:1902668
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of chemokine-mediated signaling pathway (IEA) | GO:0070100
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of epithelial cell proliferation involved in lung morphogenesis (IEA) | GO:2000795
- negative regulation of interleukin-17-mediated signaling pathway (IEA) | GO:1903882
- negative regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070108
- negative regulation of interleukin-4-mediated signaling pathway (IEA) | GO:1902215
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of retrograde transport, endosome to Golgi (IEA) | GO:1905280
- negative regulation of thyroid hormone receptor activity (IEA) | GO:1904168
- neutral lipid metabolic process (IEA) | GO:0006638
- Notch signaling pathway involved in negative regulation of venous endothelial cell fate commitment (IEA) | GO:2000796
- nucleotide-binding oligomerization domain containing 2 signaling pathway (IEA) | GO:0070431
- obsolete aerotaxis on or near other organism involved in symbiotic interaction (IEA) | GO:0052238
- obsolete aging (IEA) | GO:0007568
- obsolete AT binding (IEA) | GO:0006328
- obsolete bent DNA binding (IEA) | GO:0006326
- obsolete extracellular carbohydrate transport (IEA) | GO:0006859
- obsolete fertilization, exchange of chromosomal proteins (IEA) | GO:0035042
- obsolete induction by organism of defense-related symbiont reactive oxygen species production (IEA) | GO:0052401
- obsolete loss of asymmetric budding (IEA) | GO:0007122
- obsolete metabolism by symbiont of host xylan (IEA) | GO:0052177
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete negative regulation of adhesion of symbiont to host epithelial cell (IEA) | GO:1905227
- obsolete negative regulation of cytochrome-c oxidase activity (IEA) | GO:1905376
- obsolete negative regulation of mating-type specific transcription from RNA polymerase II promoter (IEA) | GO:0001198
- obsolete positive regulation of 1-phosphatidylinositol-3-kinase activity (IEA) | GO:0061903
- obsolete positive regulation of adhesion of symbiont to host epithelial cell (IEA) | GO:1905228
- obsolete positive regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process (IEA) | GO:1902964
- obsolete positive regulation of nucleic acid-templated transcription (IEA) | GO:1903508
- obsolete positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response (IEA) | GO:0006990
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete reproduction (IEA) | GO:0000003
- obsolete response to lipoic acid (IEA) | GO:1990479
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- ommochrome biosynthetic process (IEA) | GO:0006727
- organelle organization (IEA) | GO:0006996
- P granule disassembly (IEA) | GO:1903864
- peripheral tolerance induction (IEA) | GO:0002465
- phagocytosis (IEA) | GO:0006909
- phenylpropanoid metabolic process (IEA) | GO:0009698
- photosynthesis, light harvesting in photosystem I (IEA) | GO:0009768
- photosynthesis, light harvesting in photosystem II (IEA) | GO:0009769
- photosynthetic NADP+ reduction (IEA) | GO:0009780
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of dendrite extension (IEA) | GO:1903861
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of embryonic pattern specification (IEA) | GO:1902877
- positive regulation of inner ear receptor cell differentiation (IEA) | GO:2000982
- positive regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070109
- positive regulation of interleukin-4-mediated signaling pathway (IEA) | GO:1902216
- positive regulation of intestinal epithelial cell development (IEA) | GO:1905300
- positive regulation of mitotic sister chromatid segregation (IEA) | GO:0062033
- positive regulation of nuclear-transcribed mRNA poly(A) tail shortening (IEA) | GO:0060213
- positive regulation of protein autoubiquitination (IEA) | GO:1902499
- primary ureteric bud growth (IEA) | GO:0060682
- primitive hemopoiesis (IEA) | GO:0060215
- pronephric glomerulus morphogenesis (IEA) | GO:0035775
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- quercetin biosynthetic process (IEA) | GO:1901734
- quercetin metabolic process (IEA) | GO:1901732
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of cardiac muscle tissue regeneration (IEA) | GO:1905178
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of chemokine (C-C motif) ligand 20 production (IEA) | GO:1903884
- regulation of chondrocyte hypertrophy (IEA) | GO:1903041
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of epidermal growth factor receptor signaling pathway involved in heart process (IEA) | GO:1905282
- regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070107
- regulation of macropinocytosis (IEA) | GO:1905301
- regulation of microglial cell migration (IEA) | GO:1904139
- regulation of phospholipid catabolic process (IEA) | GO:0060696
- regulation of synaptic transmission, glycinergic (IEA) | GO:0060092
- release of sequestered calcium ion into cytosol by endoplasmic reticulum (IEA) | GO:1903514
- response to ethylene (IEA) | GO:0009723
- response to meiotic DNA replication checkpoint signaling (IEA) | GO:0072441
- response to methamphetamine hydrochloride (IEA) | GO:1904313
- response to oxygen radical (IEA) | GO:0000305
- response to reactive oxygen species (IEA) | GO:0000302
- response to singlet oxygen (IEA) | GO:0000304
- RNA polymerase III type 2 promoter transcriptional preinitiation complex assembly (IEA) | GO:0001021
- signal transduction involved in meiotic DNA replication checkpoint (IEA) | GO:0072440
- sister chromatid cohesion (IEA) | GO:0007062
- somatic diversification of immune receptors (IEA) | GO:0002200
- sperm aster formation (IEA) | GO:0035044
- sperm DNA decondensation (IEA) | GO:0035041
- spindle assembly involved in female meiosis (IEA) | GO:0007056
- spindle assembly involved in male meiosis I (IEA) | GO:0007054
- sterol regulatory element binding protein cleavage involved in ER-nuclear sterol response pathway (IEA) | GO:0006992
- sterol regulatory element binding protein import into nucleus involved in sterol depletion response (IEA) | GO:0006993
- striated muscle myosin thick filament assembly (IEA) | GO:0071688
- synaptic transmission involved in micturition (IEA) | GO:0060084
- synaptic vesicle docking (IEA) | GO:0016081
- terpenoid catabolic process (IEA) | GO:0016115
- trachea development (IEA) | GO:0060438
- transcription initiation from RNA polymerase III type 1 promoter (IEA) | GO:0001022
- triterpenoid catabolic process (IEA) | GO:0016105
- vacuolar acidification (IEA) | GO:0007035
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
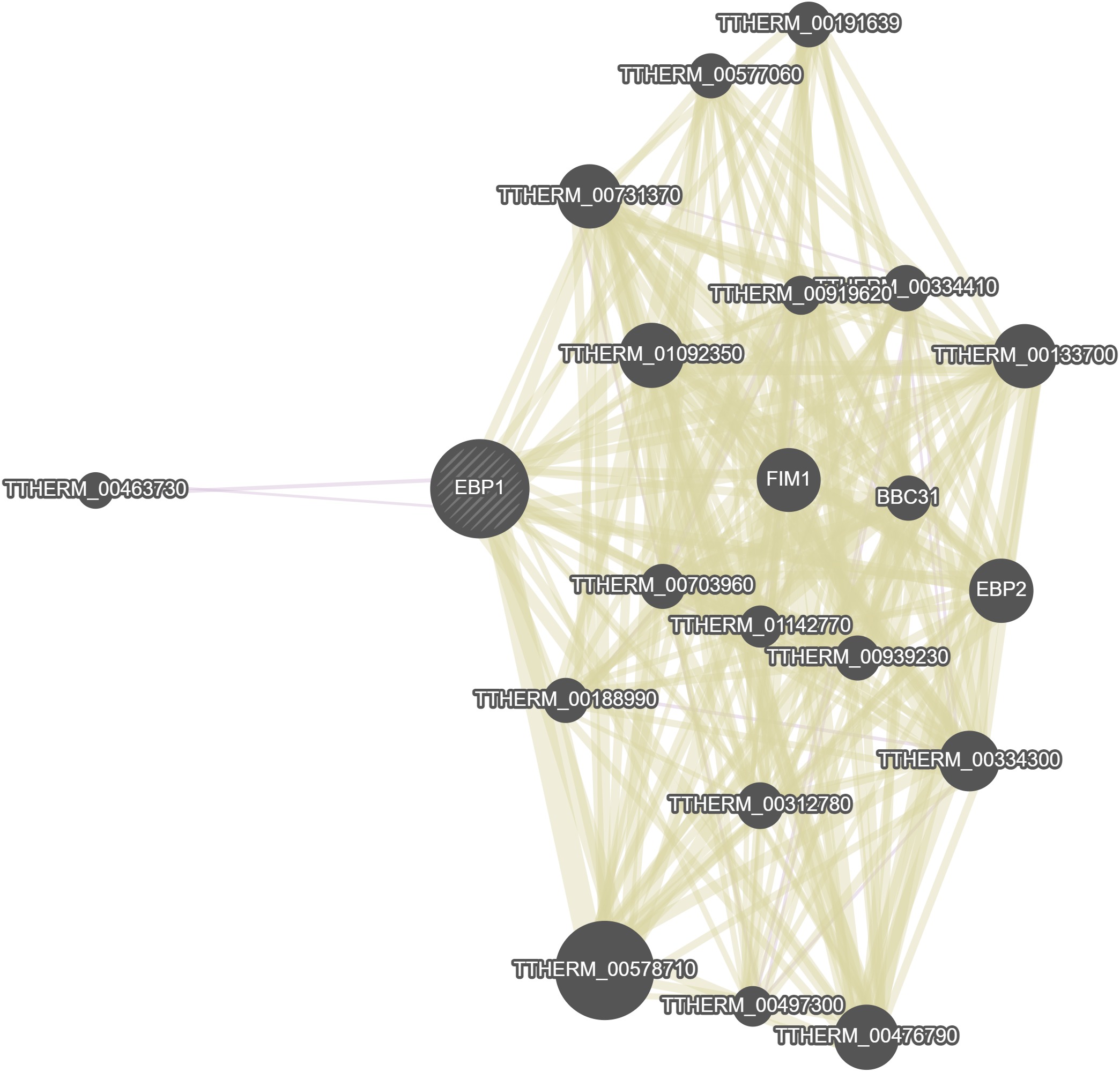
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00558460(coding)
ATGACTGAAGTTGTAACTACTGCAAATCAAAGCAATTTTTTATCTAGATAAGAAATGCTC
TAATGGATCAATGATAGTTTAAAACTTAATGAGAAGAAGATAGAAAATTTAGGCACTGGA
GCTATTTACTGTCAATTATTTGATTTGGTGTACCCTGGAAAGGTGCAGATGGTAAAGGTT
AACTGGAAAGCAAAGTATGAAAATGAATATGTTTCAAATTTCAAGATTTTATAGTAAGCA
TTTGACAAGATGAGTGTTAATAAGCCCATTGATGTGAATAAGTTAATTAAATGTAAATAC
TAAGATAATCTTGAATTTTTACAGTGGATGAAAAAATATCATGATCAATTTCGTAAAAAT
TAAGATAAGCCTTACGATGCTATAGGTAGAAGAAATGGATAAGTTTTGATTGATAGAGAA
TACCCAAATGATAAAATTCCTACTCATCCTCACTTCTAAGCAATTAAAGAATAAGCAAGC
AATATTTTAGATAACAGTGTGAATCATTCATCAAAGCATAATTTTTCAAGGCATTCTAAT
AAAAAGAGCTAAACTCATTTTGTACCTTCTGATTAAGGAAAGCAAATGAGTCATTCTTGC
TAATCTTCGATATCCAATTTATCTATTGATAATGAATTTAATCACCTAGACGAAGCAGAT
GCAGTTAAACCTTATCCATCTACTAATAAAATATAATCTGACTCTCTAATAGAAAGACTT
AAAGATGAACGAAATTTTTATTTTTCTAAGATACAGAGTATTGATTAAATGCTAGATAAT
ATTTCAGACAAGAATCCAATGACTATTAACGATATTAAAAAATATATTAAAACCATTCTA
TACAGTACATAAGATTAAATTTTAACTATATTGGATGATGGAACTGTAACTTTGGCTGAC
AAATAGCTTAATAACGTATCTATTAATTAATTTGAGTCAATGGAAGGTCTTGAGTAGCTC
AAAAAAGAAATACTCAGTACTGATTAATAAATCAAAAGTAGTGCCAAGAAAAACAAAAAG
AAAAACACAAAAACATATGATAACAATTAAGAAAATAAAAACACACTTTTAGTGCCTTAA
CATTAATAAAGCGAAACATTTTCAAATAGATCAAATTAAGGTTTTAAGACCCCAAAAAGT
AGTAAATACTCTTATGAGCAATCTCTATAGCTTGATGATTTCACCACATCAATAAATTAA
AATTCCAATAATAACTTTATAACATTTAACTAAAACAATAATAAATCTCAGTCAATACAA
TCAAATTTTGCTCATAATAAAAAAGATTCAGAACCATTATATGAAATTTTTATAAATGAA
GAATAAAATTAAAAAAATAAATAAAACGAAAATTTAAAAAACTAATCTCAAAGCTCATAT
GGAGATTTAATAAAAATACAACAAGTATCAGAAATATAATTACCAAATAACGATGTGTTT
GAACTAAATTAAAATCTTCTTGAAAGCTAAGACGAAGACTTACTAGGAAATTTAGAGTGA
>TTHERM_00558460(gene)
TCATATATTTCTGTTGCAAGCAAGCAAGCATTTGTTTTTAATTTCTATTTGAATTAACAA
AAAAATTTAAATTAATTCAATTTTTATAAAATCAATTATCAAATATAGAACTATTTAGCG
AAATACCAAACTAAATAAAAATAGCAATATAACATATTACAAAAAAAGGAAAGGCTTAAA
CAAAAGGCAATGACTGAAGTTGTAACTACTGCAAATCAAAGCAATTTTTTATCTAGATAA
GAAATGCTCTAATGGATCAATGATAGTTTAAAACTTAATGAGAAGAAGATAGAAAATTTA
GGCACTGGAGCTATTTACTGTCAATTATTTGATTTGGTGTACCCTGGAAAGGTGCAGATG
GTAAAGGTTAACTGGAAAGCAAAGTATGAAAATGAATATGTTTCAAATTTCAAGATTTTA
TAGTAAGCATTTGACAAGATGAGTGTTAATAAGCCCATTGATGTGAATAAGTTAATTAAA
TGTAAATACTAAGATAATCTTGAATTTTTACAGTGGATGAAAAAATATCATGATCAATTT
CGTAAAAATTAAGATAAGCCTTACGATGCTATAGGTAGAAGAAATGGATAAGTTTTGATT
GATAGAGAATACCCAAATGATAAAATTCCTACTCATCCTCACTTCTAAGCAATTAAAGAA
TAAGCAAGCAATATTTTAGATAACAGTGTGAATCATTCATCAAAGCATAATTTTTCAAGG
CATTCTAATAAAAAGAGCTAAACTCATTTTGTACCTTCTGATTAAGGAAAGCAAATGAGT
CATTCTTGCTAATCTTCGATATCCAATTTATCTATTGATAATGAATTTAATCACCTAGAC
GAAGCAGATGCAGTTAAACCTTATCCATCTACTAATAAAATATAATCTGACTCTCTAATA
GAAAGACTTAAAGATGAACGAAATTTTTATTTTTCTAAGATACAGAGTATTGATTAAATG
CTAGATAATATTTCAGACAAGAATCCAATGACTATTAACGATATTAAAAAATATATTAAA
ACCATTCTATACAGTACATAAGATTAAATTTTAACTATATTGGATGATGGAACTGTAACT
TTGGCTGACAAATAGCTTAATAACGTATCTATTAATTAATTTGAGTCAATGGAAGGTCTT
GAGTAGCTCAAAAAAGAAATACTCAGTACTGATTAATAAATCAAAAGTAGTGCCAAGAAA
AACAAAAAGAAAAACACAAAAACATATGATAACAATTAAGAAAATAAAAACACACTTTTA
GTGCCTTAACATTAATAAAGCGAAACATTTTCAAATAGATCAAATTAAGGTTTTAAGACC
CCAAAAAGTAGTAAATACTCTTATGAGCAATCTCTATAGCTTGATGATTTCACCACATCA
ATAAATTAAAATTCCAATAATAACTTTATAACATTTAACTAAAACAATAATAAATCTCAG
TCAATACAATCAAATTTTGCTCATAATAAAAAAGATTCAGAACCATTATATGAAATTTTT
ATAAATGAAGAATAAAATTAAAAAAATAAATAAAACGAAAATTTAAAAAACTAATCTCAA
AGCTCATATGGAGATTTAATAAAAATACAACAAGTATCAGAAATATAATTACCAAATAAC
GATGTGTTTGAACTAAATTAAAATCTTCTTGAAAGCTAAGACGAAGACTTACTAGGAAAT
TTAGAGTGAGAATCTCATTAACTCTGTATTTATGTATTTTATGTTATATTATTTAGATAA
ATAATTAACTTAAATTGCGTATCAAAACTAATTAGTTTCATATCATTTATTTCTTATTCA
TGTTAATGTAAATTTAAAATGTTAAAAAATAAATATA>TTHERM_00558460(protein)
MTEVVTTANQSNFLSRQEMLQWINDSLKLNEKKIENLGTGAIYCQLFDLVYPGKVQMVKV
NWKAKYENEYVSNFKILQQAFDKMSVNKPIDVNKLIKCKYQDNLEFLQWMKKYHDQFRKN
QDKPYDAIGRRNGQVLIDREYPNDKIPTHPHFQAIKEQASNILDNSVNHSSKHNFSRHSN
KKSQTHFVPSDQGKQMSHSCQSSISNLSIDNEFNHLDEADAVKPYPSTNKIQSDSLIERL
KDERNFYFSKIQSIDQMLDNISDKNPMTINDIKKYIKTILYSTQDQILTILDDGTVTLAD
KQLNNVSINQFESMEGLEQLKKEILSTDQQIKSSAKKNKKKNTKTYDNNQENKNTLLVPQ
HQQSETFSNRSNQGFKTPKSSKYSYEQSLQLDDFTTSINQNSNNNFITFNQNNNKSQSIQ
SNFAHNKKDSEPLYEIFINEEQNQKNKQNENLKNQSQSSYGDLIKIQQVSEIQLPNNDVF
ELNQNLLESQDEDLLGNLE