Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00561680Standard Name
RPL35 (Ribosomal Protein of the Large subunit #35)Aliases
PreTt27928 | 74.m00205 | 3720.m00095Description
RPL35 ribosomal protein L35; Homolog of Yeast RPL35, Bacterial RPL29; Large ribosomal subunit protein uL29, eukaryotesGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- ionotropic glutamate receptor complex (IEA) | GO:0008328
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 11beta-hydroxyprogesterone binding (IEA) | GO:1903879
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- CCR10 chemokine receptor binding (IEA) | GO:0031735
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete sulfur-containing amino acid secondary active transmembrane transporter activity (IEA) | GO:1901680
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- prenyltransferase activity (IEA) | GO:0004659
- prephenate dehydrogenase (NADP+) activity (IEA) | GO:0004665
- protein histidine kinase activity (IEA) | GO:0004673
- proteinase activated receptor binding (IEA) | GO:0031871
- type 2 proteinase activated receptor binding (IEA) | GO:0031873
Biological Process
- asymmetric protein localization (IEA) | GO:0008105
- basement membrane organization (IEA) | GO:0071711
- box H/ACA sno(s)RNA 3'-end processing (IEA) | GO:0000495
- calcium ion export (IEA) | GO:1901660
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to diacyl bacterial lipopeptide (IEA) | GO:0071726
- coenzyme A metabolic process (IEA) | GO:0015936
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- learning or memory (IEA) | GO:0007611
- maturation of 5.8S rRNA from tetracistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, 2S rRNA, LSU-rRNA) (IEA) | GO:0000487
- microtubule depolymerization (IEA) | GO:0007019
- mitochondrial membrane organization (IEA) | GO:0007006
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of glycoprotein metabolic process (IEA) | GO:1903019
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- negative regulation of stomatal opening (IEA) | GO:1902457
- negative regulation of telomere single strand break repair (IEA) | GO:1903824
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete autophagic death (sensu Saccharomyces) (IEA) | GO:0007577
- obsolete bent DNA binding (IEA) | GO:0006326
- obsolete larval behavior (sensu Insecta) (IEA) | GO:0007627
- obsolete MAPK cascade involved in ascospore formation (IEA) | GO:1903695
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete RING-like zinc finger domain-mediated complex assembly (IEA) | GO:0071536
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- pigment accumulation in response to UV light (IEA) | GO:0043478
- pigment accumulation in tissues in response to UV light (IEA) | GO:0043479
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of peroxisome proliferator activated receptor signaling pathway (IEA) | GO:0035360
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903691
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- pyrimidine nucleobase fermentation (IEA) | GO:0043466
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of glycoprotein metabolic process (IEA) | GO:1903018
 Domains
Domains
- ( PF00831 ) Ribosomal L29 protein
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
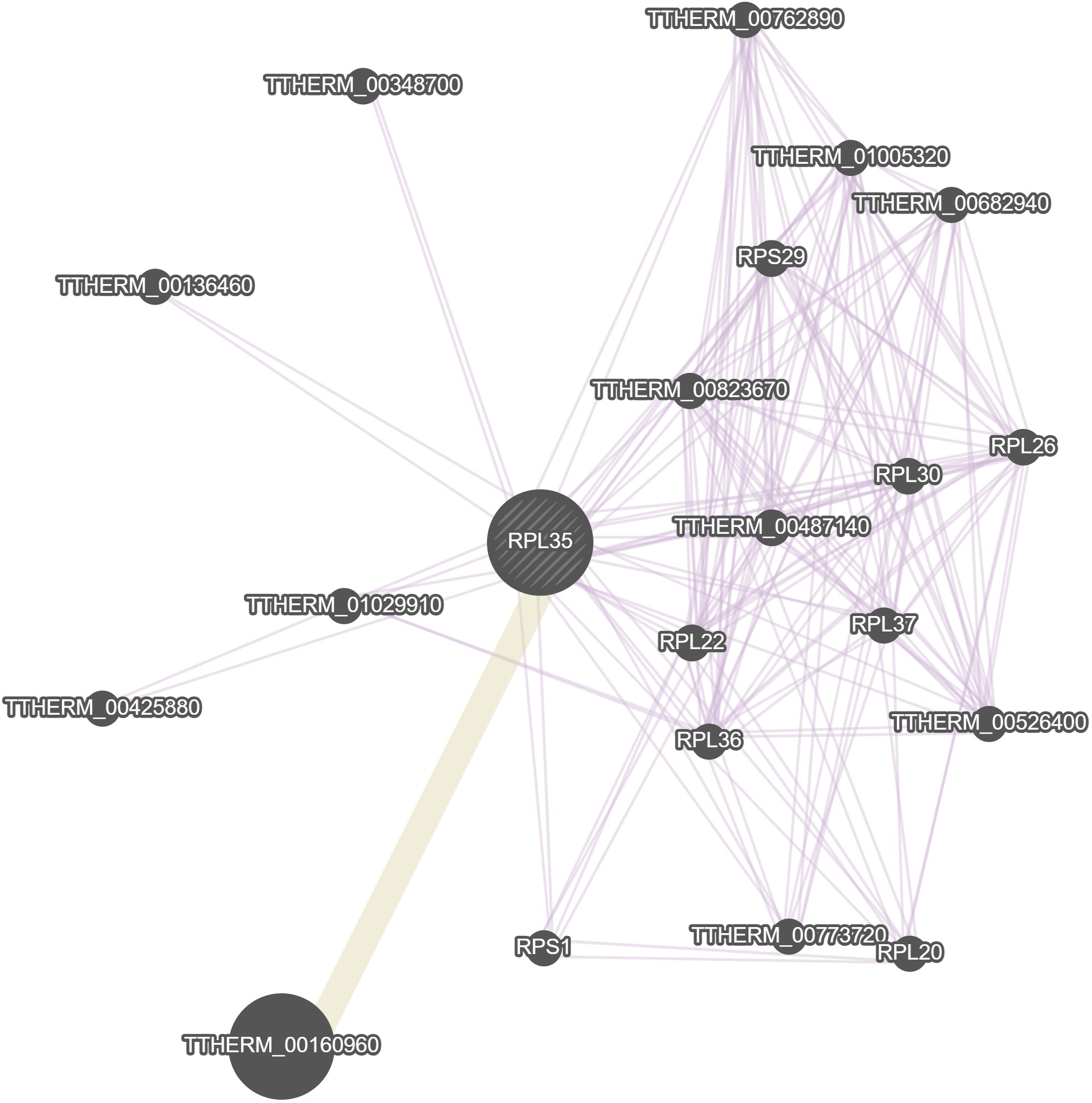
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00561680(coding)
ATGGATAAGTCTGTCAGAGTTTTCAAGCTTAGAACCTAAACTGAAGAATAGCTCGTCGGT
GAACTCGGCAAGCTCTAAACTGAACTTTCCCAACTCAGAATTGCCAAGATTGCTGGTGGT
ACTGCTAACAAGCTCGGCAGAATTGGCATCGTCAGAAAGGCTATTGCTAAATACTTGACC
ATTATCAACGAAAAGAGAAGACAAGCTGTCAAGGACTAATTCAAGGGTAAGTCTTTGAAG
CCTTTGGACATTAGAGTCAAGAAGACTAGAGCTATTAGAAGAAAACTCACCAAGAAGCAA
AGAGAAGCTGTCCTCGTCAAGACCCAAAAGAAATTAAACAACTTCGGTCTTAGAAAGTTC
GCCCTCAAGGCTTGA>TTHERM_00561680(gene)
AAATTAAATTAAAATTAATATACAAACAACAAAGTAAATAACAAGGATAAATAAATCAAA
ATGGATAAGTCTGTCAGAGTTTTCAAGCTTAGAACCTAAACTGAAGAATAGCTCGTCGGT
GAACTCGGCAAGCTCTAAACTGAACTTTCCCAACTCAGAATTGCCAAGATTGCTGGTGGT
ACTGCTAACAAGCTCGGCAGAATTGGCATCGTCAGAAAGGCTATTGCTAAATACTTGACC
ATTATCAACGAAAAGAGAAGACAAGCTGTCAAGGACTAATTCAAGGGTAAGTCTTTGAAG
CCTTTGGACATTAGAGTCAAGAAGACTAGAGCTATTAGAAGAAAACTCACCAAGAAGCAA
AGAGAAGCTGTCCTCGTCAAGACCCAAAAGAAATTAAACAACTTCGGTCTTAGAAAGTTC
GCCCTCAAGGCTTGAGTTTGAATGAAATATTTTATTTCTATTCTAATATATAAAAAATTT
AATATTATATACAAATTTTATCGAGATTGTACTTGTTTAATAACTACTTATATGCCTTAT
TTAAACGAAGAAATATTTCCTCTTAAAAACTCATCTCATTCTTCAATTAATTAATCAATC
AAAATTTATTTGTATGTATATTATTTATTTTATACTTGCTTGTGTTTTTTTAAAATCGGC
AACTTCGTATCTAATTTAATTAAT>TTHERM_00561680(protein)
MDKSVRVFKLRTQTEEQLVGELGKLQTELSQLRIAKIAGGTANKLGRIGIVRKAIAKYLT
IINEKRRQAVKDQFKGKSLKPLDIRVKKTRAIRRKLTKKQREAVLVKTQKKLNNFGLRKF
ALKA