Gene Model Identifier
TTHERM_00561799
Standard Name
TKU71
Aliases
3720.m00655
Description
TKU71 TKU70 ATP-dependent DNA helicase II 70 kDa subunit (Ku70); DNA-dependent ATP-dependent helicase; homolog to human KU70 DNA helicase and yeast YKU70; Ku70/Ku80 beta-barrel domain
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
>TTHERM_00561799(coding)
ATGGAATTTGATGAATTTGAAGATGACTTCGTCGGCGGAGACGCTGGAGAAGGTGGTGAC
CAAGCTTAGGGATTTGATTTCTAGGAAAACGAATTTATTCCTGATCTCATAAGTAAAAAA
GATGCAGTCATCTTTTTAATTGACTGCAACAAGAAGATCTTCCAAAACGAGTTTAATGAT
GAAGTTTAGTTTAAAGTAATACTAAAGAGTATTTAGAGTTTCTTCAAGTCAAAGATTATT
TCAAGTTTAGATGACTAAGTTTCTGTTATTTTCTACAATGTGAAACAAACAAATAATGAT
TTGAACTTTAAGGGGATCAATATAGTTTATAATCTTAGCACTCCTTCAGCAGAAATGATT
AAAAACGTAGTTAAGTTAGGAGATAATTTTGAAAGAGATTTCGGTTTTGCAGACAATGAA
ACACCTTTCCACGAAGCATTGTGGACTTGCAATAGCATTTTTACAAAATTAGATAGTAAA
AAATTTATAATGAGAATATTCCTATTCACCAACAATGATTAGCCCCATTCAGATAATAAA
GATTTTTTAGATAAGTGCTTTAACTAAGCAAGAACTTTGGCAGAAAAGAATATCTAAATT
GAGTTGTTCCCTCTTGCTACTAGAAACTAAAAGTTTGATATCAGAAAATTCTATTCAAGC
ATCATTACCTTTGATGTAGATGAAATTAATGAAGCTGTTATCGATACATCAGATAAAGTT
TTGGATATCCAACACAGATTACGTCAAAAAGAATACAAAAAAAGGGCAGTCAACAGATTA
CTTTTTACTTTGGGTGATGAAACTAGATTGGGAGTCAAATTGTTTTGCCCATTCGTGAAA
TAAAGACGCCCTCTTGCCAAATAATTAGAATCTAACACTAATAAACTTTTAAAAAGGACA
GTAAAGCATATTTGCAAAGAAACAGGTTAGTCTTTATATCAAAACCAAATTTCCACATGT
GTTCACATTGGAGGAGAGAAGGTTAAAGTTAGTAAAGGTGATATTAATGAAATCAAATCA
TTCGAAGAACCAGGTATGCGTTTAATAGGATTTAAACCTTCAGGAACACTTAAGGCTTAC
CACAACTATAGAACAAGCTACTTTGTTTACCCAGATGATTATCATGTAAAGGGATCATCT
CAATCTTTTCATGCTTTGATCTCATAAATGATTGCCAAAGATAAAATAGCTATTGTTAGA
TTCATTCCCCGTAAAGGCACTTAGGTTAGGTTCTGTGCTTTACTTCCATAAGAAGAAAGT
TATGATGAAGACCATGTTTAAACACCTCCAGGCTTTCATTTGATTTTCTTACCATATGCT
GATGATTAGAGAAATTTAGGTTCCATCAGACCTTAATAAAAGGAAGAAGTTAGCAGAAAT
TTACTAAATGCAGCTAAAATGATGTGCAATGCATTAGATTTAGCTAATTTTGATTTTAGA
AATTTCGAAGATCCTTCACTATAGAAATTTTTTGCTCATCTTCAGGCACATGCCTTATAA
GAAGAAAACCCAGAAAATCCAGCAGACTTGATCTAACCTGATGAAGAAGGTATGAAACGC
TGTGAAGATATAATTAAGCTATTTGAAGAAGCATTGAAGTTAGATGTATAAGATGATGAC
CAACCTCAAGCCGCTTAAAAAAAACCAAGATAAAAAAAGGTAGCTGTTGAAAAGTAACCT
GCTGCACAAAAGAAATAACCTGCTCCAAAAAAGCCTGCTTAAAACAAAAAATAAGTAAAA
AAAGAAGAAGAGTCTGAAGCAAGCAGTATAGGTGATTATGACTCTGAGGACTCCTTTATC
GATAATGAAGAAGATGAAATTAAGCCTAAAAAAAGAGGAGTTTAAAACAAAAAAGTGACT
AATTAAAGAAAAGTTAATAAAAAAGTTGAATACAGTGATGAAGATGATGAGATAAGCTAT
GAAGAAAAACCAAAACGAAAAGCAGCAGGGAAGAAACAAATGAGCGAAGAAGAAAAACTT
ATTTCACATGTAGAAGCTAATTTAAAAGAAGAATTCAACCCAGATGGTTTGACAATATCT
GAGCTTAAATAGTATCTTAAGTTCAAAGGACTTAAAAATGTTGGTAAAAAGGAAGAATTG
GTTGATACCATTTTAAGACACTTAGAAAGAAATTGA
>TTHERM_00561799(gene)
ATTAATCATCAATCAATCATAGCTGTTGAAAATACAAATCTTTTTTCAGCATAAATAAAA
GATTCAAGCAAGACAAACAATATCGTTTTAATTAAATGGAATTTGATGAATTTGAAGATG
ACTTCGTCGGCGGAGACGCTGGAGAAGGTGGTGACCAAGCTTAGGGATTTGATTTCTAGG
AAAACGAATTTATTCCTGATCTCATAAGTAAAAAAGATGCAGTCATCTTTTTAATTGACT
GCAACAAGAAGATCTTCCAAAACGAGTTTAATGATGAAGTTTAGTTTAAAGTAATACTAA
AGAGTATTTAGAGTTTCTTCAAGTCAAAGATTATTTCAAGTTTAGATGACTAAGTTTCTG
TTATTTTCTACAATGTGAAACAAACAAATAATGATTTGAACTTTAAGGGGATCAATATAG
TTTATAATCTTAGCACTCCTTCAGCAGAAATGATTAAAAACGTAGTTAAGTTAGGAGATA
ATTTTGAAAGAGATTTCGGTTTTGCAGACAATGAAACACCTTTCCACGAAGCATTGTGGA
CTTGCAATAGCATTTTTACAAAATTAGATAGTAAAAAATTTATAATGAGAATATTCCTAT
TCACCAACAATGATTAGCCCCATTCAGATAATAAAGATTTTTTAGATAAGTGCTTTAACT
AAGCAAGAACTTTGGCAGAAAAGAATATCTAAATTGAGTTGTTCCCTCTTGCTACTAGAA
ACTAAAAGTTTGATATCAGAAAATTCTATTCAAGCATCATTACCTTTGATGTAGATGAAA
TTAATGAAGCTGTTATCGATACATCAGATAAAGTTTTGGATATCCAACACAGATTACGTC
AAAAAGAATACAAAAAAAGGGCAGTCAACAGATTACTTTTTACTTTGGGTGATGAAACTA
GATTGGGAGTCAAATTGTTTTGCCCATTCGTGAAATAAAGACGCCCTCTTGCCAAATAAT
TAGAATCTAACACTAATAAACTTTTAAAAAGGACAGTAAAGCATATTTGCAAAGAAACAG
GTTAGTCTTTATATCAAAACCAAATTTCCACATGTGTTCACATTGGAGGAGAGAAGGTTA
AAGTTAGTAAAGGTGATATTAATGAAATCAAATCATTCGAAGAACCAGGTATGCGTTTAA
TAGGATTTAAACCTTCAGGAACACTTAAGGCTTACCACAACTATAGAACAAGCTACTTTG
TTTACCCAGATGATTATCATGTAAAGGGATCATCTCAATCTTTTCATGCTTTGATCTCAT
AAATGATTGCCAAAGATAAAATAGCTATTGTTAGATTCATTCCCCGTAAAGGCACTTAGG
TTAGGTTCTGTGCTTTACTTCCATAAGAAGAAAGTTATGATGAAGACCATGTTTAAACAC
CTCCAGGCTTTCATTTGATTTTCTTACCATATGCTGATGATTAGAGAAATTTAGGTTCCA
TCAGACCTTAATAAAAGGAAGAAGTTAGCAGAAATTTACTAAATGCAGCTAAAATGATGT
GCAATGCATTAGATTTAGCTAATTTTGATTTTAGAAATTTCGAAGATCCTTCACTATAGA
AATTTTTTGCTCATCTTCAGGCACATGCCTTATAAGAAGAAAACCCAGAAAATCCAGCAG
ACTTGATCTAACCTGATGAAGAAGGTATGAAACGCTGTGAAGATATAATTAAGCTATTTG
AAGAAGCATTGAAGTTAGATGTATAAGATGATGACCAACCTCAAGCCGCTTAAAAAAAAC
CAAGATAAAAAAAGGTAGCTGTTGAAAAGTAACCTGCTGCACAAAAGAAATAACCTGCTC
CAAAAAAGCCTGCTTAAAACAAAAAATAAGTAAAAAAAGAAGAAGAGTCTGAAGCAAGCA
GTATAGGTGATTATGACTCTGAGGACTCCTTTATCGATAATGAAGAAGATGAAATTAAGC
CTAAAAAAAGAGGAGTTTAAAACAAAAAAGTGACTAATTAAAGAAAAGTTAATAAAAAAG
TTGAATACAGTGATGAAGATGATGAGATAAGCTATGAAGAAAAACCAAAACGAAAAGCAG
CAGGGAAGAAACAAATGAGCGAAGAAGAAAAACTTATTTCACATGTAGAAGCTAATTTAA
AAGAAGAATTCAACCCAGATGGTTTGACAATATCTGAGCTTAAATAGTATCTTAAGTTCA
AAGGACTTAAAAATGTTGGTAAAAAGGAAGAATTGGTTGATACCATTTTAAGACACTTAG
AAAGAAATTGAATGAAATTTTAATATTTAATAAAACTTGTAATTAACTAATTATAAATTA
TTTTCTTGTAATTGTATTTATTTTATTCTGAAAGCAATTCAATTGTTTTCTTTTTATTCA
AATAAATCCTAATAAGGAATTCTCTTCATTA
>TTHERM_00561799(protein)
MEFDEFEDDFVGGDAGEGGDQAQGFDFQENEFIPDLISKKDAVIFLIDCNKKIFQNEFND
EVQFKVILKSIQSFFKSKIISSLDDQVSVIFYNVKQTNNDLNFKGINIVYNLSTPSAEMI
KNVVKLGDNFERDFGFADNETPFHEALWTCNSIFTKLDSKKFIMRIFLFTNNDQPHSDNK
DFLDKCFNQARTLAEKNIQIELFPLATRNQKFDIRKFYSSIITFDVDEINEAVIDTSDKV
LDIQHRLRQKEYKKRAVNRLLFTLGDETRLGVKLFCPFVKQRRPLAKQLESNTNKLLKRT
VKHICKETGQSLYQNQISTCVHIGGEKVKVSKGDINEIKSFEEPGMRLIGFKPSGTLKAY
HNYRTSYFVYPDDYHVKGSSQSFHALISQMIAKDKIAIVRFIPRKGTQVRFCALLPQEES
YDEDHVQTPPGFHLIFLPYADDQRNLGSIRPQQKEEVSRNLLNAAKMMCNALDLANFDFR
NFEDPSLQKFFAHLQAHALQEENPENPADLIQPDEEGMKRCEDIIKLFEEALKLDVQDDD
QPQAAQKKPRQKKVAVEKQPAAQKKQPAPKKPAQNKKQVKKEEESEASSIGDYDSEDSFI
DNEEDEIKPKKRGVQNKKVTNQRKVNKKVEYSDEDDEISYEEKPKRKAAGKKQMSEEEKL
ISHVEANLKEEFNPDGLTISELKQYLKFKGLKNVGKKEELVDTILRHLERN
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
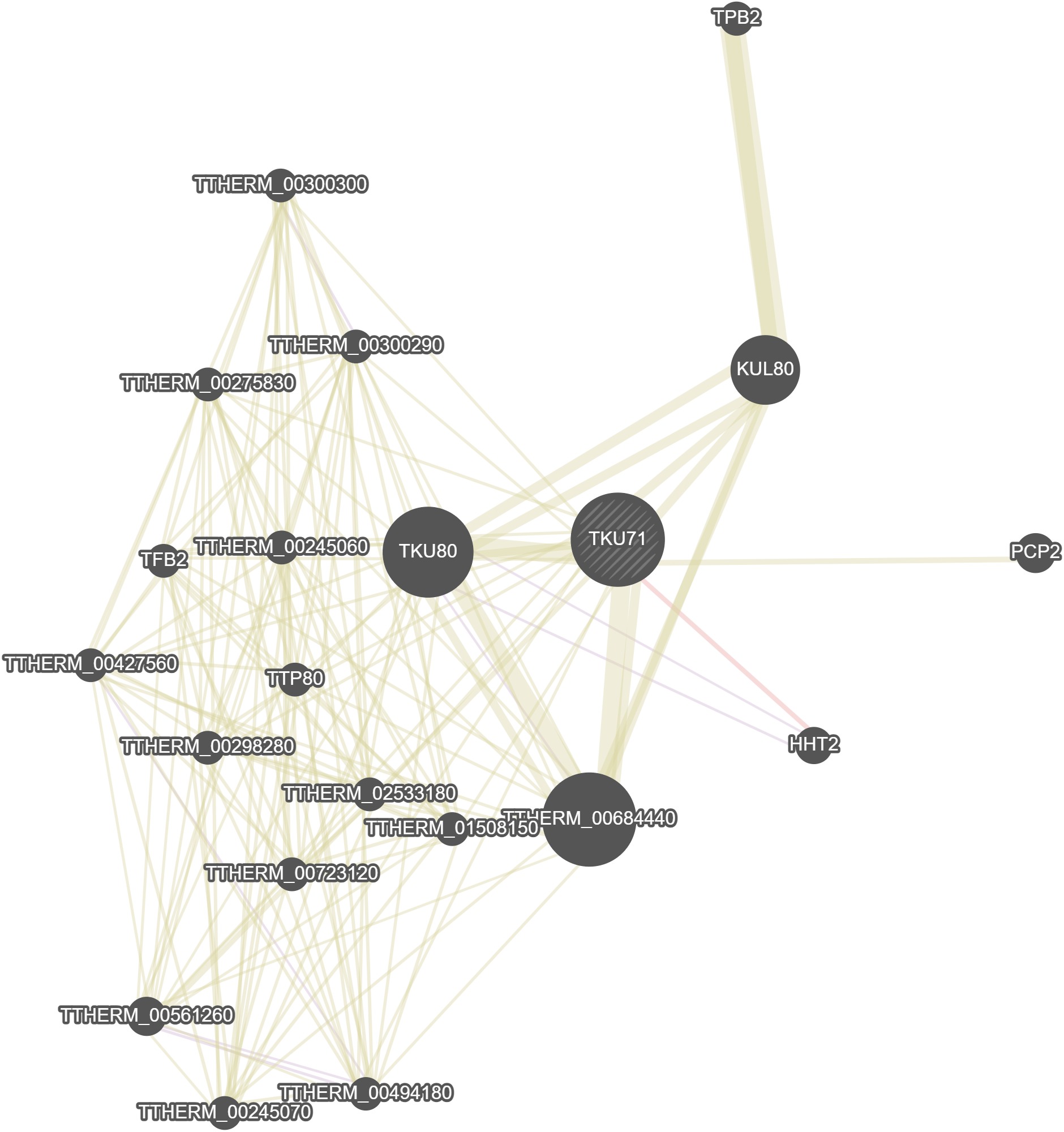
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences