Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00568050Standard Name
HMGB6 (High mobility group box protein 6)Aliases
PreTt10434 | 76.m00136 | 3810.m01998Description
HMGB6 high mobility group (HMG)-box protein; HMG box family protein; Upstream Binding Factor 1Genome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- homodimeric serine palmitoyltransferase complex (IEA) | GO:0002179
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- obsolete axonemal dynein heavy chain (IEA) | GO:0001683
- obsolete axonemal dynein intermediate chain (IEA) | GO:0001684
- obsolete axonemal dynein intermediate light chain (IEA) | GO:0001685
- obsolete ectoplasm (IEA) | GO:0043265
- SAM complex (IEA) | GO:0001401
- subrhabdomeral cisterna (IEA) | GO:0016029
Molecular Function
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-keto-3-deoxy-L-rhamnonate aldolase activity (IEA) | GO:0106099
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- all-trans retinol 3,4-desaturase activity (IEA) | GO:0061896
- benomyl transporter activity (IEA) | GO:0015242
- dolichyl pyrophosphate Glc2Man9GlcNAc2 alpha-1,2-glucosyltransferase activity (IEA) | GO:0106073
- ferric enterobactin:proton symporter activity (IEA) | GO:0015345
- ferric triacetylfusarinine C:proton symporter activity (IEA) | GO:0015346
- lipopolysaccharide heptosyltransferase activity (IEA) | GO:0008920
- metarhodopsin binding (IEA) | GO:0016030
- obsolete amiloride transmembrane transporter activity (IEA) | GO:0015240
- obsolete calcium-dependent secreted phospholipase A2 activity (IEA) | GO:0004625
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete globin (IEA) | GO:0001524
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete zinc, iron permease activity (IEA) | GO:0015342
- oligoxyloglucan reducing-end-specific cellobiohydrolase activity (IEA) | GO:0033945
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase (ATP) activity (IEA) | GO:0004612
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- phosphoglycerate dehydrogenase activity (IEA) | GO:0004617
- phosphoribosyl-AMP cyclohydrolase activity (IEA) | GO:0004635
- proteinase activated receptor binding (IEA) | GO:0031871
- siderophore uptake transmembrane transporter activity (IEA) | GO:0015344
- siderophore-iron transmembrane transporter activity (IEA) | GO:0015343
- Swi5-Sfr1 complex binding (IEA) | GO:1905334
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
Biological Process
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- basement membrane organization (IEA) | GO:0071711
- cell fate determination (IEA) | GO:0001709
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to dopamine (IEA) | GO:1903351
- cellular response to homocysteine (IEA) | GO:1905375
- coenzyme A biosynthetic process (IEA) | GO:0015937
- coenzyme A metabolic process (IEA) | GO:0015936
- defense response to tumor cell (IEA) | GO:0002357
- DNA replication preinitiation complex assembly (IEA) | GO:0071163
- embryo implantation (IEA) | GO:0007566
- epithelium development (IEA) | GO:0060429
- female pregnancy (IEA) | GO:0007565
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- gamma-delta T cell lineage commitment (IEA) | GO:0002365
- glycerophosphodiester transmembrane transport (IEA) | GO:0001407
- glycolytic process from glycerol (IEA) | GO:0061613
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- inactivation of recombination (HML) (IEA) | GO:0007537
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- internal genitalia morphogenesis (IEA) | GO:0035260
- leukocyte chemotaxis involved in immune response (IEA) | GO:0002233
- lung connective tissue development (IEA) | GO:0060427
- lung epithelium development (IEA) | GO:0060428
- lung pattern specification process (IEA) | GO:0060432
- lung saccule development (IEA) | GO:0060430
- maltohexaose transport (IEA) | GO:2001103
- medium-chain fatty acid transport (IEA) | GO:0001579
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of carotenoid biosynthetic process (IEA) | GO:1904142
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete activation of MAPKK during sporulation (sensu Saccharomyces) (IEA) | GO:0000204
- obsolete age dependent accumulation of genetic damage (IEA) | GO:0007570
- obsolete age dependent increased protein content (IEA) | GO:0007573
- obsolete age-dependent general metabolic decline involved in chronological cell aging (IEA) | GO:0001323
- obsolete aging (IEA) | GO:0007568
- obsolete cell aging (IEA) | GO:0007569
- obsolete cell aging (sensu Saccharomyces) (IEA) | GO:0007574
- obsolete MAPK cascade involved in ascospore formation (IEA) | GO:1903695
- obsolete negative regulation of cytochrome-c oxidase activity (IEA) | GO:1905376
- obsolete negative regulation of leucine-tRNA ligase activity (IEA) | GO:1903634
- obsolete nucleolar fragmentation (IEA) | GO:0007576
- obsolete nucleolar size increase (IEA) | GO:0007575
- obsolete regulation of cell growth by detection of nuclear:cytoplasmic ratio (IEA) | GO:0001559
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete reproduction (IEA) | GO:0000003
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- obsolete sodium/potassium transport (IEA) | GO:0006815
- obsolete synthesis of RNA primer involved in premeiotic DNA replication (IEA) | GO:1902980
- ommochrome biosynthetic process (IEA) | GO:0006727
- parturition (IEA) | GO:0007567
- phagocytosis (IEA) | GO:0006909
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of chemokine (C-C motif) ligand 20 production (IEA) | GO:1903886
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of response to water deprivation (IEA) | GO:1902584
- primary lung bud formation (IEA) | GO:0060431
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- pyrimidine deoxyribonucleotide metabolic process (IEA) | GO:0009219
- pyrimidine nucleotide salvage (IEA) | GO:0032262
- pyrimidine ribonucleotide metabolic process (IEA) | GO:0009218
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of attachment of mitotic spindle microtubules to kinetochore (IEA) | GO:1902423
- regulation of cell growth (IEA) | GO:0001558
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of chitin-based cuticle tanning (IEA) | GO:0007564
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of ecdysteroid secretion (IEA) | GO:0007555
- regulation of gastric motility (IEA) | GO:1905333
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of microglial cell migration (IEA) | GO:1904139
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of protein localization to presynapse (IEA) | GO:1905384
- regulation of RNA binding transcription factor activity (IEA) | GO:1905255
- sesquiterpenoid biosynthetic process (IEA) | GO:0016106
- sodium-independent prostaglandin transport (IEA) | GO:0071720
- sperm aster formation (IEA) | GO:0035044
- sperm DNA decondensation (IEA) | GO:0035041
- striated muscle myosin thick filament assembly (IEA) | GO:0071688
- synaptic vesicle docking (IEA) | GO:0016081
- synaptic vesicle priming (IEA) | GO:0016082
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- triterpenoid catabolic process (IEA) | GO:0016105
- tRNA import into mitochondrion (IEA) | GO:0016031
- vesicle coating (IEA) | GO:0006901
 Domains
Domains
- ( PF00505 ) HMG (high mobility group) box
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
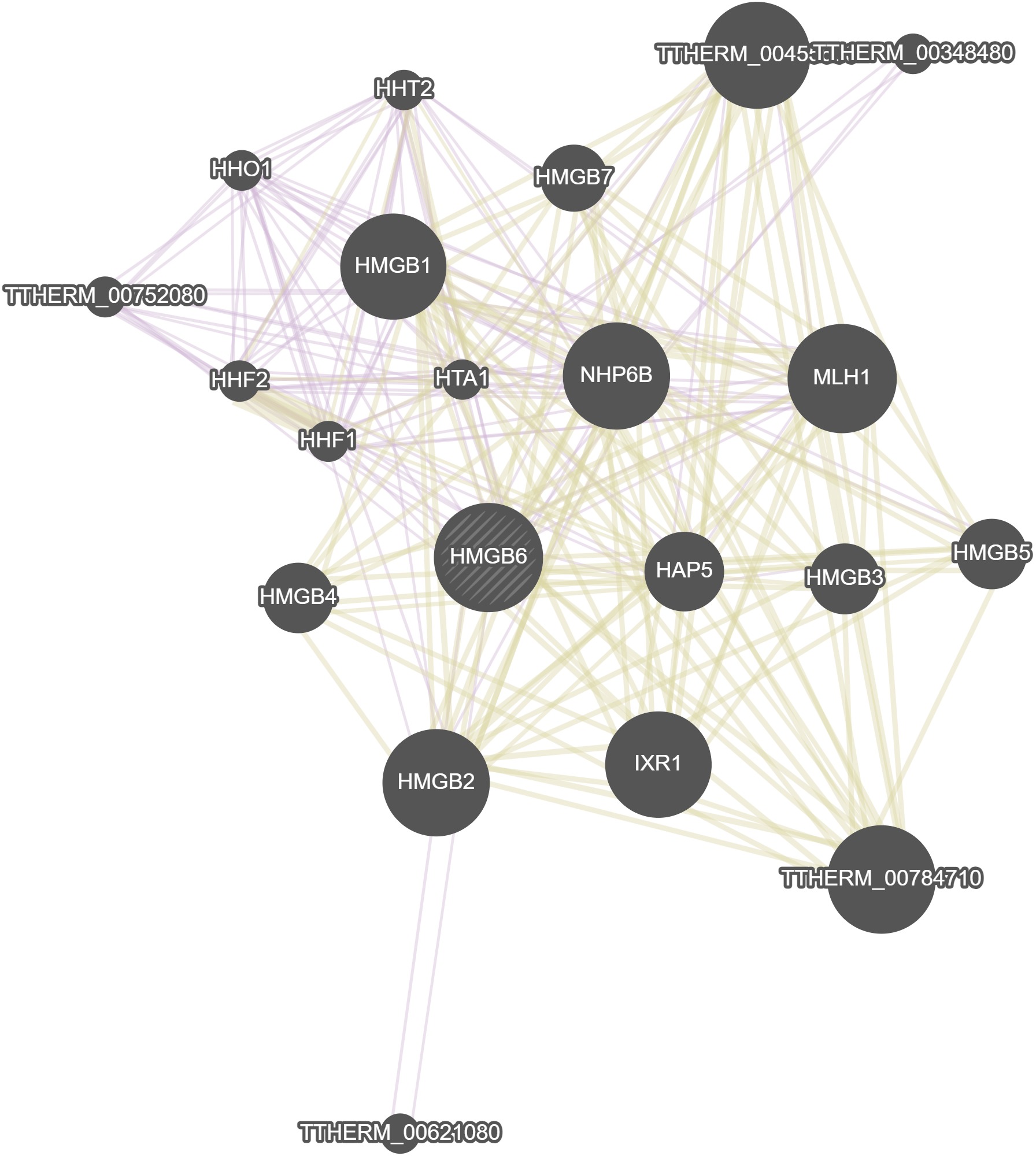
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:31932604: Nabeel-Shah S, Ashraf K, Saettone A, Garg J, Derynck J, Lambert JP, Pearlman RE, Fillingham J (2020) Nucleus-specific linker histones Hho1 and Mlh1 form distinct protein interactions during growth, starvation and development in Tetrahymena thermophila. Scientific reports 10(1):168
 Sequences
Sequences
>TTHERM_00568050(coding)
ATGCCAGGCAATGATGATGATGATGGATTTGATGACTTCATTAATTAATTGAAGGAAAAA
AAGGAGCATTTAAAAGTGAAATTATAGGGAGGAAGCGAAGAAATTCGCAAGCCCAAAAAG
CCATTAACTATCTTTTTAAGATTCCACATGGAGAAGTTTAATTAAATAAAAATGAAAAAT
CAAGATTGGACACCTAATATGATAACTCAATATTTAAAGAAGCAATGGGAGAGCATGAGT
GAGGCTTAAAAAGAAAGATATATCTAGACCTACGAATAGGATTTTAAAAAGTATAATAAG
TTAATTGACCTTTACAACAAGCTCACTCTTAAACCTAAGAAACCTATTACTCCTTATTTT
TAATTTTTTTCTGAAAATATTGAAGTATATTTAAAGAATTACCCTGAAAAATCGCATAAT
GAAATCACTAAATTGATTGGAGATGATTGGAATAATTTTGATTCCAAAAAAAAAAGTGAG
ATGAAAAAAGATTATGAAAAATAGAAGCAACTTTATGAAGTCTAGTTACAAAAATTTAAG
TAAAGAGAAAATAACTATATAAAATATGATGACTATAATCAAAAACTAGAAGATTATGAA
AAGGAGTTAGAAGAAGAATTAGATAATATCCGTTCTAAATTCCCAAAAGTTGCATCAACC
ATTCCAAAAAAGAAAAATTAGAAATAGAATTCAAAAGATAAATAAAATTTAAAGGATGAA
AAACAACAATAGAAGGATGAAAGCGAACAAGAATAGGAAGATCTTGAAGTGAAAATTGAA
GAAGAAGACGAACAAGGTTTAGATAAAGAAGATGAGGATGAAGGATAAGAAGAAGAAGAA
AATTTATCAAATGAAAGTGATTAAAAAAGAAATAAAAGCAAATCAAAAAGTAAGAAAGTT
TAAAAGGAGACCTAAAAAGCTTCAAATAATAAAAGAGAGTAAAAAGAGTAAAAGTTAAAA
GAAAAATCAGATAATAAAATAGATAAAGCCATTAAAGCAGCATAAGAAGAGGGCAGATCA
GGATTAAGAAAGAGGGAAGCAACTAAATAAACTAAAACTTAAGAAAATGAAAATAAATAA
GTAGAAAGAAGTGTATCTAAATCCTTAAAAACAAGATCATCTTCTAAAAAAGCATAAGAG
CATACTAAAATGAAAAGATCAAGATCTTAAATTGCACTTAAACAGTAATAAGAAAAAGAG
GAATCAAGTAAATAAGTGCCATAAGTAGCTAAAAGATCATCATCTTAATAGTCTTAAAAT
GTTACTTCTAAAGATAGTTAACAAAAAAAAACAAAGAATTAGGAAGCATCCAGATCTTCA
ACTAGAATTAAAGAGTAGAAGCTCAATGTAAAGAAAGAAGCATCAAAAAAGGTTGAAAAT
GATAAAAAGAAAGATGAAAAGAAAAAAGAAGTTTAAAGAGAAAGTTTAAAAAATATAAAA
ATCTTAAAAAGAAAGAATAAAGATAGAGATGCACAAGATGATGAAGTAGAAGAAGATATC
GAAGAGACTGTTCAAAAAAAATAAAAACTTGTTCAAGAAAAAATTATCACACACATATTA
GGAATGAGAGAATATAGAGGCAAAGTTAAATATTTATTAAGATTGAAAAATGATGAGTAA
TACTGGGCTAAGAGAGATGATCCTATTTTTAACGAAAATAAAAACCAATAAAAGTTGTCT
GTATTCTTAAAGTAAATTAAAAAACAAAAAAAGTGA>TTHERM_00568050(gene)
CTTTTAAATTTCAAATCTTAAAATATTAAAAAAAAGAGTGGGAAAATAAAGAGGTAGAAA
CAATTTATAGAGTAACACAATATTGAAAAAATGCCAGGCAATGATGATGATGATGGATTT
GATGACTTCATTAATTAATTGAAGGAAAAAAAGGAGCATTTAAAAGTGAAATTATAGGGA
GGAAGCGAAGAAATTCGCAAGCCCAAAAAGCCATTAACTATCTTTTTAAGATTCCACATG
GAGAAGTTTAATTAAATAAAAATGAAAAATCAAGATTGGACACCTAATATGATAACTCAA
TATTTAAAGAAGCAATGGGAGAGCATGAGTGAGGCTTAAAAAGAAAGATATATCTAGACC
TACGAATAGGATTTTAAAAAGTATAATAAGTTAATTGACCTTTACAACAAGCTCACTCTT
AAACCTAAGAAACCTATTACTCCTTATTTTTAATTTTTTTCTGAAAATATTGAAGTATAT
TTAAAGAATTACCCTGAAAAATCGCATAATGAAATCACTAAATTGATTGGAGATGATTGG
AATAATTTTGATTCCAAAAAAAAAAGTGAGATGAAAAAAGATTATGAAAAATAGAAGCAA
CTTTATGAAGTCTAGTTACAAAAATTTAAGTAAAGAGAAAATAACTATATAAAATATGAT
GACTATAATCAAAAACTAGAAGATTATGAAAAGGAGTTAGAAGAAGAATTAGATAATATC
CGTTCTAAATTCCCAAAAGTTGCATCAACCATTCCAAAAAAGAAAAATTAGAAATAGAAT
TCAAAAGATAAATAAAATTTAAAGGATGAAAAACAACAATAGAAGGATGAAAGCGAACAA
GAATAGGAAGATCTTGAAGTGAAAATTGAAGAAGAAGACGAACAAGGTTTAGATAAAGAA
GATGAGGATGAAGGATAAGAAGAAGAAGAAAATTTATCAAATGAAAGTGATTAAAAAAGA
AATAAAAGCAAATCAAAAAGTAAGAAAGTTTAAAAGGAGACCTAAAAAGCTTCAAATAAT
AAAAGAGAGTAAAAAGAGTAAAAGTTAAAAGAAAAATCAGATAATAAAATAGATAAAGCC
ATTAAAGCAGCATAAGAAGAGGGCAGATCAGGATTAAGAAAGAGGGAAGCAACTAAATAA
ACTAAAACTTAAGAAAATGAAAATAAATAAGTAGAAAGAAGTGTATCTAAATCCTTAAAA
ACAAGATCATCTTCTAAAAAAGCATAAGAGCATACTAAAATGAAAAGATCAAGATCTTAA
ATTGCACTTAAACAGTAATAAGAAAAAGAGGAATCAAGTAAATAAGTGCCATAAGTAGCT
AAAAGATCATCATCTTAATAGTCTTAAAATGTTACTTCTAAAGATAGTTAACAAAAAAAA
ACAAAGAATTAGGAAGCATCCAGATCTTCAACTAGAATTAAAGAGTAGAAGCTCAATGTA
AAGAAAGAAGCATCAAAAAAGGTTGAAAATGATAAAAAGAAAGATGAAAAGAAAAAAGAA
GTTTAAAGAGAAAGTTTAAAAAATATAAAAATCTTAAAAAGAAAGAATAAAGATAGAGAT
GCACAAGATGATGAAGTAGAAGAAGATATCGAAGAGACTGTTCAAAAAAAATAAAAACTT
GTTCAAGAAAAAATTATCACACACATATTAGGAATGAGAGAATATAGAGGCAAAGTTAAA
TATTTATTAAGATTGAAAAATGATGAGTAATACTGGGCTAAGAGAGATGATCCTATTTTT
AACGAAAATAAAAACCAATAAAAGTTGTCTGTATTCTTAAAGTAAATTAAAAAACAAAAA
AAGTGAAACAACCTTCGTTCCTTATTCAAAAAAAAAAATCTAGCAATAAATTAATAAATT
CTAAGAACTACTTATTGATTGACTTTTCTAAAGAGATATTTTAGAAAAGAATATTTAATA
TAATATTGATATAGGTAATTTATCATAAATAAATAAAATAAGTAAGGAATGTATAAATCA
ATCAATAAGTTCATGTAAATAAATCTATGGTTTATAAATT>TTHERM_00568050(protein)
MPGNDDDDGFDDFINQLKEKKEHLKVKLQGGSEEIRKPKKPLTIFLRFHMEKFNQIKMKN
QDWTPNMITQYLKKQWESMSEAQKERYIQTYEQDFKKYNKLIDLYNKLTLKPKKPITPYF
QFFSENIEVYLKNYPEKSHNEITKLIGDDWNNFDSKKKSEMKKDYEKQKQLYEVQLQKFK
QRENNYIKYDDYNQKLEDYEKELEEELDNIRSKFPKVASTIPKKKNQKQNSKDKQNLKDE
KQQQKDESEQEQEDLEVKIEEEDEQGLDKEDEDEGQEEEENLSNESDQKRNKSKSKSKKV
QKETQKASNNKREQKEQKLKEKSDNKIDKAIKAAQEEGRSGLRKREATKQTKTQENENKQ
VERSVSKSLKTRSSSKKAQEHTKMKRSRSQIALKQQQEKEESSKQVPQVAKRSSSQQSQN
VTSKDSQQKKTKNQEASRSSTRIKEQKLNVKKEASKKVENDKKKDEKKKEVQRESLKNIK
ILKRKNKDRDAQDDEVEEDIEETVQKKQKLVQEKIITHILGMREYRGKVKYLLRLKNDEQ
YWAKRDDPIFNENKNQQKLSVFLKQIKKQKK