Gene Model Identifier
TTHERM_00578560
Standard Name
FAP91
(Flagellar Associated Protein 91)
Aliases
PreTt05749 | 78.m00121 | 3733.m00016
Description
FAP91 MAATS1 putative; Calmodulin and spoke-associated complex (CSC) protein; homolog of the Chlamydomonas CSC protein FAP91; Cilia- and flagella-associated protein 91
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
>TTHERM_00578560(coding)
ATGGCCACAACAACTAATATTCTTCATGACGTTGTTAGAGATTAGTCAATGGTAAATTAT
GTCTCAAATAGAGATAGACCACTCTATTTTAAGAGGCCTCTTGTGCCTCAAATGACAGAC
ATTCCTTTGCATATCTCTCGTCCTCCTGTAGATTAATAAGTGGATTATTAAACTATGTAG
ATGCAATAAAATGCTACCATAGTTGAGGCACCTACAAAAGATGCTTTTGTATAAACAGAT
TATAGAGAAAGTGAGACTTAAACTGATCCTTATACTCCTAAATGTTTTGTTCGTGATGGA
GATCATCCTGAGGTTATGGAATTAAAAGATTATAAGTATGGAAAGGGACTCCCTGCATCA
ATTGAAGAGTTAGAATAAATTGAATTAAATCGTGAAAAAGTGTGGTTTGAAAATTCCTTG
CCTCCCATTTCTGATGAAGCATCGTTTAATTTACGTCGTAAATTGATGGAAGAATAAGAA
CTACGTGAATGGAGCAAAAAAGAAAATGAAATAAAAAAATTCTAAAATGAGAAACTTTAT
TTGCTCCAATAGGCTTTAATTGAAAGAGAAAAAGAAGTCGAGGACAAGAGCTAAGAAAGA
ATAGAAGAAATCAGGTAATAAAAGACTGAGCACAAGAATAGATAGATTGCTAAGATTTAA
AGAAAAAAAATTAAAATTGACAGAAAGATGACTAAAAGCAGAAAAATGCAGGGTAAAGAG
ACTTTGAAGCGTGATATTATTGAAGATTATGCTAATTTTGCTTCTAGAGTTTACGCTGGT
ATTACTCACGAAGGTCTTTCTCTTGATAAGATAGCTAACAAATATGAAGTATAACCTCTA
GCATTAGGAAACTATGAAATGCTTCAAGCCCTCCATGAAGGTATTCGCCCAAGAGAGTTT
GAAACAAGAGTCAATCTCAAAAAGGAAATTAAGGAAATTGAAAAGAACTATACAAGATTG
GAAAATTATCATAGAGGCGAGCTTAAAAAGGCATAAGATGAAATTAATGGAGTTCATGAA
TAAAGCAAAGCCTAATAAAAATAAGAAGGAAACAACTTTTCTTACAGAAACTTCGAAAAC
AAAATTCGTCCTGCAACTCCTACTTGGAAATATGACACTGATTTTAACATTAGTCCTTCT
CTAATAACAGAAATTTAGGAGTACAGAGGTCCTAATGGTGTTTAATTGATGCAAGCTGCT
GATAAGAGGGAAGAAGCTGTTCTCCTTTTACAAAGATTGTTAAGAGGTAGAACTACTTAG
AATATAATGTACGAAGGTAAGAAAAAGCGTACTGCCCTTATTGAAGAATTGCTTACAGTA
GCTCAAATCGAAAACTTAGAGGAAGATAAAGCTGAAGAAGTGCTCATGTAACAACATGAA
GAAAAAGTTAAGAATGCTGTACTAGAAAGCATTTAAGGAGAAGTTATAGCTGAAACTATG
GATGAATTATCAAAGGAATTGCTTCGTATCAAGCAAGAAAGAAAGATATAGCAAATGGTT
GAAATAGCTGAAAAGGACAGAAGAATAAGAGAAATCGAGGAAGCTGGTAAAAGACAAGCA
GAAGAGATTCTTAGAGACCGTGAAGACGTTCTTCACAATTAACTTATCAGAGTTCATCAA
GGCACCGTAGATACATATTTAGATTGGTTGATGTCGAATGCTTTAGAACAATCTTCTGCA
AGACAAGCCACAATAATGACAAATTTAAGAAAGGCAAAATTCAACTTGCCTTTGGAGGAC
TTCGAAAGAAAATATAATAACAATCAAACTCTTATAAAGGATCTTGTACACTCATTCCTA
ATTCCAAACGTACAACGCTCCAAGCTTAAAAAATAAATTCAATTAGAGGAAAAGAGATTT
AGCGAGGCTGCTAAGAGATCTCTTCAACAAACAATAACCCGTGCATCAAATAAGCACGCA
AATGTTTAAAATTGA
>TTHERM_00578560(gene)
AGGAAGAAGTTAAAGGAATATCAATTAACATTAATTTAGGAATATTTACCAATTATTATA
AAATGGCCACAACAACTAATATTCTTCATGACGTTGTTAGAGATTAGTCAATGGTAAATT
ATGTCTCAAATAGAGATAGACCACTCTATTTTAAGAGGCCTCTTGTGCCTCAAATGACAG
ACATTCCTTTGCATATCTCTCGTCCTCCTGTAGATTAATAAGTGGATTATTAAACTATGT
AGATGCAATAAAATGCTACCATAGTTGAGGCACCTACAAAAGATGCTTTTGTATAAACAG
ATTATAGAGAAAGTGAGACTTAAACTGATCCTTATACTCCTAAATGTTTTGTTCGTGATG
GAGATCATCCTGAGGTTATGGAATTAAAAGATTATAAGTATGGAAAGGGACTCCCTGCAT
CAATTGAAGAGTTAGAATAAATTGAATTAAATCGTGAAAAAGTGTGGTTTGAAAATTCCT
TGCCTCCCATTTCTGATGAAGCATCGTTTAATTTACGTCGTAAATTGATGGAAGAATAAG
AACTACGTGAATGGAGCAAAAAAGAAAATGAAATAAAAAAATTCTAAAATGAGAAACTTT
ATTTGCTCCAATAGGCTTTAATTGAAAGAGAAAAAGAAGTCGAGGACAAGAGCTAAGAAA
GAATAGAAGAAATCAGGTAATAAAAGACTGAGCACAAGAATAGATAGATTGCTAAGATTT
AAAGAAAAAAAATTAAAATTGACAGAAAGATGACTAAAAGCAGAAAAATGCAGGGTAAAG
AGACTTTGAAGCGTGATATTATTGAAGATTATGCTAATTTTGCTTCTAGAGTTTACGCTG
GTATTACTCACGAAGGTCTTTCTCTTGATAAGATAGCTAACAAATATGAAGTATAACCTC
TAGCATTAGGAAACTATGAAATGCTTCAAGCCCTCCATGAAGGTATTCGCCCAAGAGAGT
TTGAAACAAGAGTCAATCTCAAAAAGGAAATTAAGGAAATTGAAAAGAACTATACAAGAT
TGGAAAATTATCATAGAGGCGAGCTTAAAAAGGCATAAGATGAAATTAATGGAGTTCATG
AATAAAGCAAAGCCTAATAAAAATAAGAAGGAAACAACTTTTCTTACAGAAACTTCGAAA
ACAAAATTCGTCCTGCAACTCCTACTTGGAAATATGACACTGATTTTAACATTAGTCCTT
CTCTAATAACAGAAATTTAGGAGTACAGAGGTCCTAATGGTGTTTAATTGATGCAAGCTG
CTGATAAGAGGGAAGAAGCTGTTCTCCTTTTACAAAGATTGTTAAGAGGTAGAACTACTT
AGAATATAATGTACGAAGGTAAGAAAAAGCGTACTGCCCTTATTGAAGAATTGCTTACAG
TAGCTCAAATCGAAAACTTAGAGGAAGATAAAGCTGAAGAAGTGCTCATGTAACAACATG
AAGAAAAAGTTAAGAATGCTGTACTAGAAAGCATTTAAGGAGAAGTTATAGCTGAAACTA
TGGATGAATTATCAAAGGAATTGCTTCGTATCAAGCAAGAAAGAAAGATATAGCAAATGG
TTGAAATAGCTGAAAAGGACAGAAGAATAAGAGAAATCGAGGAAGCTGGTAAAAGACAAG
CAGAAGAGATTCTTAGAGACCGTGAAGACGTTCTTCACAATTAACTTATCAGAGTTCATC
AAGGCACCGTAGATACATATTTAGATTGGTTGATGTCGAATGCTTTAGAACAATCTTCTG
CAAGACAAGCCACAATAATGACAAATTTAAGAAAGGCAAAATTCAACTTGCCTTTGGAGG
ACTTCGAAAGAAAATATAATAACAATCAAACTCTTATAAAGGATCTTGTACACTCATTCC
TAATTCCAAACGTACAACGCTCCAAGCTTAAAAAATAAATTCAATTAGAGGAAAAGAGAT
TTAGCGAGGCTGCTAAGAGATCTCTTCAACAAACAATAACCCGTGCATCAAATAAGCACG
CAAATGTTTAAAATTGAATTTAATAATGATACTTGTTCTAGGGATAAAATGTAATTAATA
ATATCTACTAGTTTTAATGACAATAAATAAGTAAAAGAAAAGATATACATTTTCTTAATT
ATTTTAAGTTTTTAAAGAGGAGTAAGTAACCAACAAACCAATCTATCAATCAACCAATAA
TAAATAGACCCAACTATTATTAAATAAATAAATAATGATAAAATAATCAATTCTATAATC
TTAAATTCAAATATATACATGCCTGCCTACCTACCTACATATATTAATAAATTTATTGAT
TTCTATAATATTACATTTATCTCACTAAATTAATAAGTCAATAATAGTAATTTAATATGG
AATTTTCTGTTTATGTACATAAGCATCTAATTTTTCTTAGACATTTA
>TTHERM_00578560(protein)
MATTTNILHDVVRDQSMVNYVSNRDRPLYFKRPLVPQMTDIPLHISRPPVDQQVDYQTMQ
MQQNATIVEAPTKDAFVQTDYRESETQTDPYTPKCFVRDGDHPEVMELKDYKYGKGLPAS
IEELEQIELNREKVWFENSLPPISDEASFNLRRKLMEEQELREWSKKENEIKKFQNEKLY
LLQQALIEREKEVEDKSQERIEEIRQQKTEHKNRQIAKIQRKKIKIDRKMTKSRKMQGKE
TLKRDIIEDYANFASRVYAGITHEGLSLDKIANKYEVQPLALGNYEMLQALHEGIRPREF
ETRVNLKKEIKEIEKNYTRLENYHRGELKKAQDEINGVHEQSKAQQKQEGNNFSYRNFEN
KIRPATPTWKYDTDFNISPSLITEIQEYRGPNGVQLMQAADKREEAVLLLQRLLRGRTTQ
NIMYEGKKKRTALIEELLTVAQIENLEEDKAEEVLMQQHEEKVKNAVLESIQGEVIAETM
DELSKELLRIKQERKIQQMVEIAEKDRRIREIEEAGKRQAEEILRDREDVLHNQLIRVHQ
GTVDTYLDWLMSNALEQSSARQATIMTNLRKAKFNLPLEDFERKYNNNQTLIKDLVHSFL
IPNVQRSKLKKQIQLEEKRFSEAAKRSLQQTITRASNKHANVQN
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
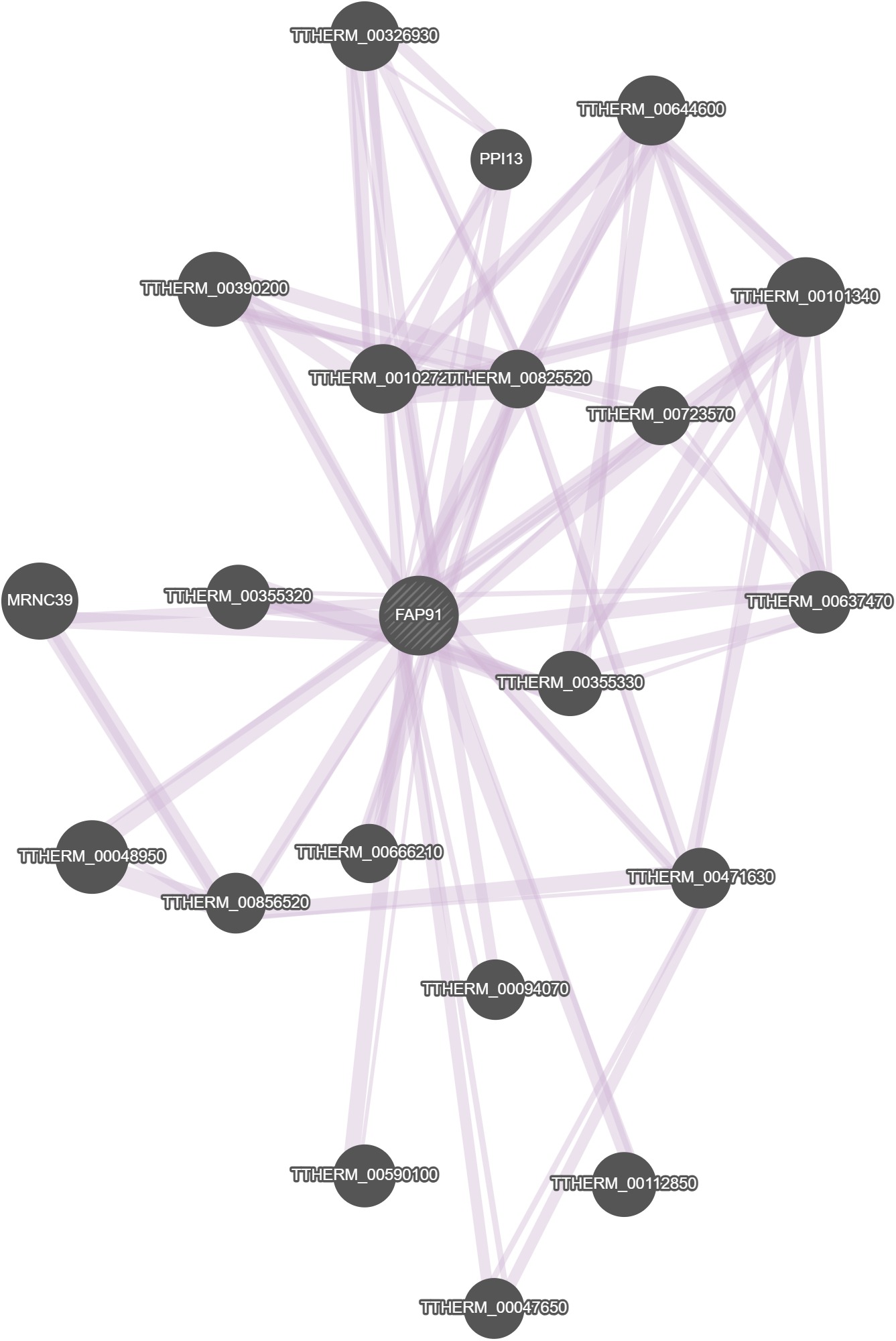
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences