Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00614790Standard Name
ATG18 (AuTophaGy 18)Aliases
PreTt19127 | 85.m00120 | 3725.m00013Description
ATG18 WD repeat protein putative; Autophagy protein; involved in programmed nuclear degradation; PROPPINGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- Myo2p-Vac17p-Vac8p transport complex (IEA) | GO:0071563
- nucleoplasmic THO complex (IEA) | GO:0000446
- small-subunit processome (IEA) | GO:0032040
- THO complex (IEA) | GO:0000347
- transcription export complex (IEA) | GO:0000346
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- ADP phosphatase activity (IEA) | GO:0043262
- axon guidance receptor activity (IEA) | GO:0008046
- cyclic-guanylate-specific phosphodiesterase activity (IEA) | GO:0071111
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- histone H3K14 deacetylase activity, NAD-dependent (IEA) | GO:0032041
- obsolete endogenous peptide receptor activity (IEA) | GO:0008529
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoric diester hydrolase activity (IEA) | GO:0008081
- RNA 2',3'-cyclic 3'-phosphodiesterase activity (IEA) | GO:0008664
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
Biological Process
- autophagy (ISS) | GO:0006914
- bacillithiol biosynthetic process (IEA) | GO:0071793
- basement membrane organization (IEA) | GO:0071711
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- commissural neuron axon guidance (IEA) | GO:0071679
- DNA recombinase mediator complex assembly (IEA) | GO:1903871
- formation of quadruple SL/U4/U5/U6 snRNP (IEA) | GO:0000353
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- intraciliary anterograde transport (IEA) | GO:0035720
- macrophage derived foam cell differentiation (IEA) | GO:0010742
- mammary gland lobule development (IEA) | GO:0061377
- mitochondrial membrane organization (IEA) | GO:0007006
- mRNA branch site recognition (IEA) | GO:0000348
- negative regulation of chemokine-mediated signaling pathway (IEA) | GO:0070100
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phospholipid translocation (IEA) | GO:0061093
- negative regulation of signal transduction (IEA) | GO:0009968
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- negative regulation of VCP-NPL4-UFD1 AAA ATPase complex assembly (IEA) | GO:1904240
- nephron tubule epithelial cell differentiation (IEA) | GO:0072160
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete activation of pro-apoptotic gene products (IEA) | GO:0008633
- obsolete induction by virus of host cell-cell fusion (IEA) | GO:0006948
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete multi-organism cellular localization (IEA) | GO:1902581
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- obsolete urea cycle intermediate metabolic process (IEA) | GO:0000051
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- phenanthrene catabolic process via trans-9(R),10(R)-dihydrodiolphenanthrene (IEA) | GO:0018956
- photosynthetic NADP+ reduction (IEA) | GO:0009780
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of peroxisome proliferator activated receptor signaling pathway (IEA) | GO:0035360
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903691
- priming of cellular response to stress (IEA) | GO:0080136
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- prostate induction (IEA) | GO:0060514
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of antisense RNA transcription (IEA) | GO:0060194
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of dendrite extension (IEA) | GO:1903859
- regulation of L-arginine import into cell (IEA) | GO:1902826
- regulation of sprouting angiogenesis (IEA) | GO:1903670
- regulation of transcription from RNA polymerase II promoter by galactose (IEA) | GO:0000431
- response to camptothecin (IEA) | GO:1901563
- response to paraquat (IEA) | GO:1901562
- single-organism intracellular transport (IEA) | GO:1902582
- steroid biosynthetic process (IEA) | GO:0006694
- striated muscle contraction (IEA) | GO:0006941
- syncytium formation (IEA) | GO:0006949
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
 Domains
Domains
- ( PF21032 ) PROPPIN
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
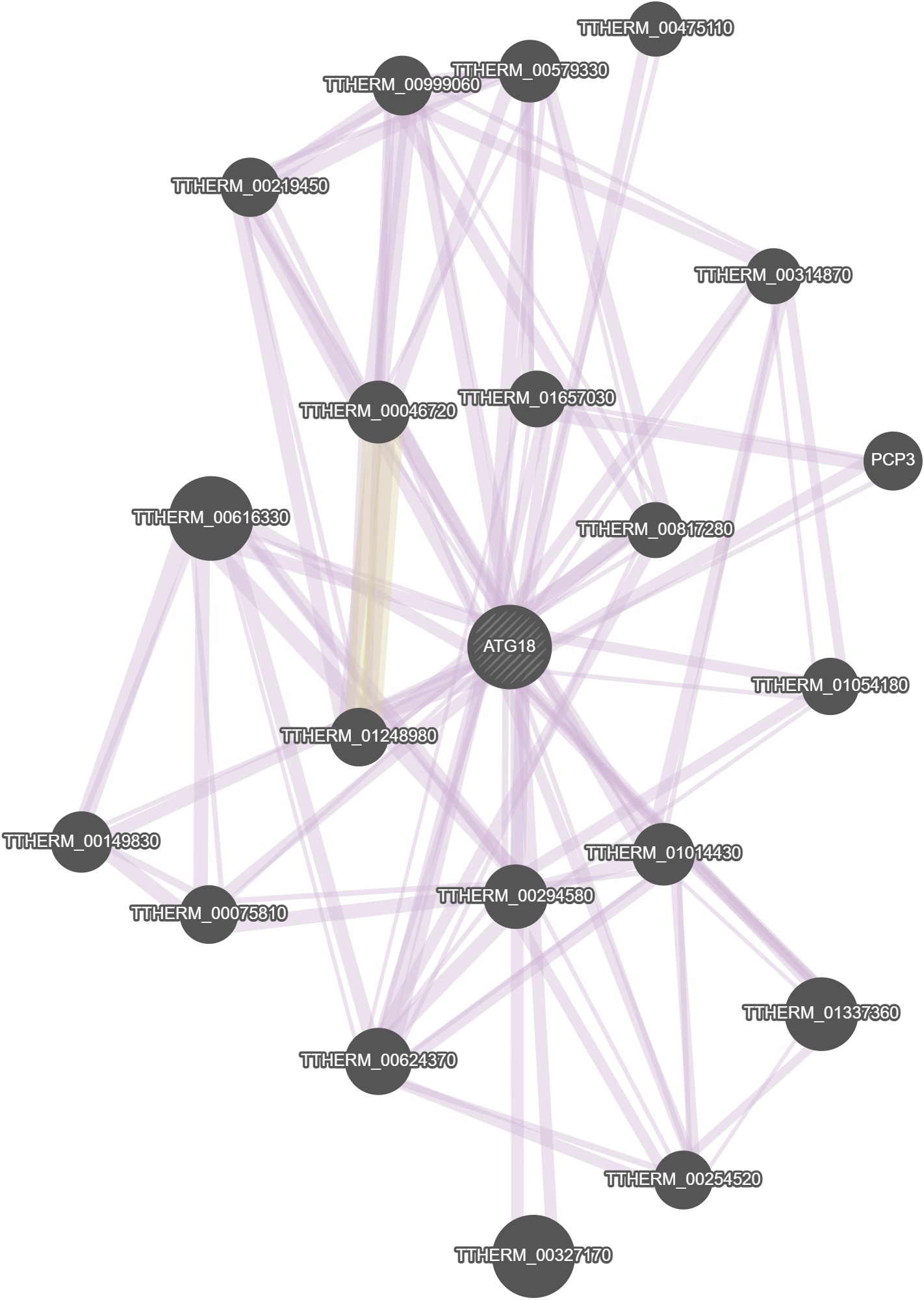
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:22366125: Liu ML, Yao MC (2012) Role of ATG8 and autophagy in programmed nuclear degradation in Tetrahymena thermophila. Eukaryotic cell 11(4):494-506
 Sequences
Sequences
>TTHERM_00614790(coding)
ATGCAAAAGTAAAATATAGTTTCAGGCTTTGAAGTCATAGATAACTTTTCGGATGATGAA
ACACATCCTAATAAATAGCTGAACTAGCCTAGATAATAAAAATCTGATATAAAAAGCTCT
AAATAGCAATCAAAATAAGATGAAAACATTACAAAATAAGAGTAGAAAAATAAAATAGAA
TAGTAATAGCAATTCATAGAAAATTCATAAAACTACGAATAGACAGATGAAACAGATGAC
ACTGAATTTCTGCATAAAAATATTCCAAAGATATATGCAATTACATTTTCGTAAACAGGA
AAAACTGTAGCAATAGGAACTGAAGAAGGGTTGAGAATATAATCATCTGAATCATTAAAA
GTTGGAATGTTAAATAAAAAATATATTATGGAAAAAGAAAAAAACTCTTTTTATTGTGGT
ATTAAGCTTGTTTCTTTAATGTTTGATACTTATATTTGTGCTTTAGTCACTGATAATAAG
AACAAATAATATCCTGATTCTAAAGTTTATATTTGGGATTAAAAAAATAGTCAAATAATA
GGTTAGTATGAATTCTATTCGCCAATTCTATAAATTTGTTATATGAGAGAATTCCTTTTA
GTTGCAAGCGAGTAACGTTTAGTCTTGTATGACATAGTGCAAAAAAAACCACTAAAATAT
ATTTTAGATATATCTACTCCTTAAAAATTTGTAGCTAGCTAACGTTATGACACATTCACC
TTTTGTTATTTAACAAAAAATAAAGGAATGATTATAGCAACAAATGTTTGGTATGACAAC
CCAGTTTAAATTGACTTTAATTAGGAAATAGGCGTTTCTACACCATAATCTTTAGAATAA
AGAGAATTTGATATTTCAAATATGGTTTTAAGTCATGATGGAAGTCTATTAGCTGCCTGC
GAAAAAGGTGGTAATATTATCAAAATTTTTGATACAGAAAAAGGTAAACTCAAATGTACT
CTTTACAGAGGTAAATATCCTAAAATAATTACAGACATGTAATTTAAGAAGGACTTAACT
GCTTTTGGATTAATAAGTACTTATGAATAGAAAAGTGAAGATGAAGGCACTATACATATA
TATGATATATCTATGATAAGTTAATTTAATAAGGAGCCAAATAAAATACCTTTTACTAGG
TTTTAAAATATCAGAAGCTAATAATTCAATTTCGATATTCTCAAAAAATACTCTTATGCA
TCAAAAGATACAAAATTCTAAGGGCCTTTAATTTAATTTGATCCTTAAGGTGGTAATTCA
TTTACTATTTATTCAGAATCTGATTATACACTAAGTTAGGAATATAATTTAACTTCTGAT
GAGCCTAAAAAAAGACTTACAGATAATTAAAAGCAAGATGGATGGATAGATCCTATTTAA
GATGGGTGA>TTHERM_00614790(gene)
TATTATTGAAATTGTTTAGAATTTAATATCAGAAAATCTTCATCAAAACTTAAAAAATAT
TAGCACAAAATAAATAGTAATGCAAAAGTAAAATATAGTTTCAGGCTTTGAAGTCATAGA
TAACTTTTCGGATGATGAAACACATCCTAATAAATAGCTGAACTAGCCTAGATAATAAAA
ATCTGATATAAAAAGCTCTAAATAGCAATCAAAATAAGATGAAAACATTACAAAATAAGA
GTAGAAAAATAAAATAGAATAGTAATAGCAATTCATAGAAAATTCATAAAACTACGAATA
GACAGATGAAACAGATGACACTGAATTTCTGCATAAAAATATTCCAAAGATATATGCAAT
TACATTTTCGTAAACAGGAAAAACTGTAGCAATAGGAACTGAAGAAGGGTTGAGAATATA
ATCATCTGAATCATTAAAAGTTGGAATGTTAAATAAAAAATATATTATGGAAAAAGAAAA
AAACTCTTTTTATTGTGGTATTAAGCTTGTTTCTTTAATGTTTGATACTTATATTTGTGC
TTTAGTCACTGATAATAAGAACAAATAATATCCTGATTCTAAAGTTTATATTTGGGATTA
AAAAAATAGTCAAATAATAGGTTAGTATGAATTCTATTCGCCAATTCTATAAATTTGTTA
TATGAGAGAATTCCTTTTAGTTGCAAGCGAGTAACGTTTAGTCTTGTATGACATAGTGCA
AAAAAAACCACTAAAATATATTTTAGATATATCTACTCCTTAAAAATTTGTAGCTAGCTA
ACGTTATGACACATTCACCTTTTGTTATTTAACAAAAAATAAAGGAATGATTATAGCAAC
AAATGTTTGGTATGACAACCCAGTTTAAATTGACTTTAATTAGGAAATAGGCGTTTCTAC
ACCATAATCTTTAGAATAAAGAGAATTTGATATTTCAAATATGGTTTTAAGTCATGATGG
AAGTCTATTAGCTGCCTGCGAAAAAGGTGGTAATATTATCAAAATTTTTGATACAGAAAA
AGGTAAACTCAAATGTACTCTTTACAGAGGTAAATATCCTAAAATAATTACAGACATGTA
ATTTAAGAAGGACTTAACTGCTTTTGGATTAATAAGTACTTATGAATAGAAAAGTGAAGA
TGAAGGCACTATACATATATATGATATATCTATGATAAGTTAATTTAATAAGGAGCCAAA
TAAAATACCTTTTACTAGGTTTTAAAATATCAGAAGCTAATAATTCAATTTCGATATTCT
CAAAAAATACTCTTATGCATCAAAAGATACAAAATTCTAAGGGCCTTTAATTTAATTTGA
TCCTTAAGGTGGTAATTCATTTACTATTTATTCAGAATCTGATTATACACTAAGTTAGGA
ATATAATTTAACTTCTGATGAGCCTAAAAAAAGACTTACAGATAATTAAAAGCAAGATGG
ATGGATAGATCCTATTTAAGATGGGTGATAAATTAATTACTTTACTAATTTAAAATTAAA
TTTTATAATATTAATTAATTAAATTATATTATTATATGTAATAAATAAATAAATAACTTA
ACTTATCAACGATCTTGTCTTTTTGTAACAATTAAAATAAATAATCTAAAATTTTTTTGT
TTTTACGAT>TTHERM_00614790(protein)
MQKQNIVSGFEVIDNFSDDETHPNKQLNQPRQQKSDIKSSKQQSKQDENITKQEQKNKIE
QQQQFIENSQNYEQTDETDDTEFLHKNIPKIYAITFSQTGKTVAIGTEEGLRIQSSESLK
VGMLNKKYIMEKEKNSFYCGIKLVSLMFDTYICALVTDNKNKQYPDSKVYIWDQKNSQII
GQYEFYSPILQICYMREFLLVASEQRLVLYDIVQKKPLKYILDISTPQKFVASQRYDTFT
FCYLTKNKGMIIATNVWYDNPVQIDFNQEIGVSTPQSLEQREFDISNMVLSHDGSLLAAC
EKGGNIIKIFDTEKGKLKCTLYRGKYPKIITDMQFKKDLTAFGLISTYEQKSEDEGTIHI
YDISMISQFNKEPNKIPFTRFQNIRSQQFNFDILKKYSYASKDTKFQGPLIQFDPQGGNS
FTIYSESDYTLSQEYNLTSDEPKKRLTDNQKQDGWIDPIQDG