Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00633170Standard Name
GPR37 (Gene Protein coupled Receptor 37)Aliases
PreTt27986 | 90.m00150 | GPCR37Description
GPR37 7 transmembrane receptor (secretin family) protein; G protein coupled receptor; demonstrates chemorepellent receptor activity in the presence of conditioned supernatant factor; GCR1-cAMP receptorGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- alpha6-beta1 integrin-CD151 complex (IEA) | GO:0071059
- CD19-Vav-PIK3R1 complex (IEA) | GO:0070024
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- myosin II complex (IEA) | GO:0016460
- obsolete chromosome segregation-directing complex (IEA) | GO:0043913
- obsolete ectoplasm (IEA) | GO:0043265
- obsolete intrinsic component of synaptic vesicle membrane (IEA) | GO:0098563
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-octaprenyl-6-methoxyphenol hydroxylase activity (IEA) | GO:0008681
- 3-hydroxybutyrate dehydrogenase activity (IEA) | GO:0003858
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- ABC-type amino acid transporter activity (IEA) | GO:0015424
- ABC-type D-xylose transporter activity (IEA) | GO:0015614
- ABC-type maltose transporter activity (IEA) | GO:0015423
- ABC-type oligosaccharide transporter activity (IEA) | GO:0015422
- amino acid transmembrane transporter activity (IEA) | GO:0015171
- aminotriazole transporter activity (IEA) | GO:0015241
- arginine transmembrane transporter activity (IEA) | GO:0015181
- arsenate reductase (glutaredoxin) activity (IEA) | GO:0008794
- cadmium ion transmembrane transporter activity (IEA) | GO:0015086
- carboxylic ester hydrolase activity (IEA) | GO:0052689
- carboxymethylenebutenolidase activity (IEA) | GO:0008806
- cobalamin transporter activity (IEA) | GO:0015235
- Cys-tRNA(Pro) hydrolase activity (IEA) | GO:0043907
- cytochrome o ubiquinol oxidase activity (IEA) | GO:0008827
- cytosine-purine permease activity (IEA) | GO:0015392
- drug transmembrane transporter activity (IEA) | GO:0015238
- endochitinase activity (IEA) | GO:0008843
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- galactoside 2-alpha-L-fucosyltransferase activity (IEA) | GO:0008107
- glycine transmembrane transporter activity (IEA) | GO:0015187
- high-affinity copper ion transmembrane transporter activity (IEA) | GO:0015089
- high-affinity sodium:dicarboxylate symporter activity (IEA) | GO:0015362
- iron-chelate-transporting ATPase activity (IEA) | GO:0015623
- L-aspartate transmembrane transporter activity (IEA) | GO:0015183
- L-cystine transmembrane transporter activity (IEA) | GO:0015184
- L-isoleucine transmembrane transporter activity (IEA) | GO:0015188
- lipopolysaccharide heptosyltransferase activity (IEA) | GO:0008920
- mercury (II) reductase (NADP+) activity (IEA) | GO:0016152
- monoatomic anion:monoatomic cation symporter activity (IEA) | GO:0015296
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete D-lysine oxidase activity (IEA) | GO:0043912
- obsolete negative transcription elongation factor activity (IEA) | GO:0008148
- obsolete phosphatidylinositol 3-kinase, class I, catalyst activity (IEA) | GO:0015072
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete zinc, iron permease activity (IEA) | GO:0015342
- orexigenic neuropeptide QRFP receptor binding (IEA) | GO:0031854
- pantothenate transmembrane transporter activity (IEA) | GO:0015233
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- polyamine transmembrane transporter activity (IEA) | GO:0015203
- pristanoyl-CoA oxidase activity (IEA) | GO:0016402
- purinergic nucleotide receptor activity (IEA) | GO:0001614
- pyrimidine nucleotide-sugar transmembrane transporter activity (IEA) | GO:0015165
- Ser(Gly)-tRNA(Ala) hydrolase activity (IEA) | GO:0043908
- sodium ion transmembrane transporter activity (IEA) | GO:0015081
- type 1B serotonin receptor binding (IEA) | GO:0031822
- uridine:proton symporter activity (IEA) | GO:0015394
Biological Process
- aminotriazole transport (IEA) | GO:0015899
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cytoplasmic actin-based contraction involved in forward cell motility (IEA) | GO:0060328
- D-tagatose 6-phosphate catabolic process (IEA) | GO:2001059
- DNA recombinase mediator complex assembly (IEA) | GO:1903871
- fucose metabolic process (IEA) | GO:0006004
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- glycolytic process from glycerol (IEA) | GO:0061613
- glycolytic process through fructose-6-phosphate (IEA) | GO:0061615
- GTP synthesis coupled proton transport (IEA) | GO:0015987
- hair cycle process (IEA) | GO:0022405
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- internal genitalia morphogenesis (IEA) | GO:0035260
- laminaritriose transport (IEA) | GO:2001097
- lung connective tissue development (IEA) | GO:0060427
- lung saccule development (IEA) | GO:0060430
- malolactic fermentation (IEA) | GO:0043464
- maltoheptaose catabolic process (IEA) | GO:2001123
- maltoheptaose metabolic process (IEA) | GO:2001122
- maltotetraose transport (IEA) | GO:2001099
- meiotic DNA double-strand break formation involved in meiotic gene conversion (IEA) | GO:0010781
- meiotic DNA double-strand break formation involved in reciprocal meiotic recombination (IEA) | GO:0010780
- mercury ion transport (IEA) | GO:0015694
- microtubule cytoskeleton organization involved in mitosis (IEA) | GO:1902850
- mitochondrial RNA surveillance (IEA) | GO:2000827
- mitotic nuclear division (IEA) | GO:0007067
- multicellular organism development (IEA) | GO:0007275
- N-acetylglucosamine catabolic process (IEA) | GO:0006046
- N,N'-diacetylchitobiose import (IEA) | GO:1902815
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of cellular response to oxidopamine (IEA) | GO:1905847
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of hemopoiesis (IEA) | GO:1903707
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- nitrite transport (IEA) | GO:0015707
- nurse cell nucleus anchoring (IEA) | GO:0007302
- obsolete carbamate resistance (IEA) | GO:0008161
- obsolete cyclodiene resistance (IEA) | GO:0008162
- obsolete fertilization, exchange of chromosomal proteins (IEA) | GO:0035042
- obsolete histone H4-K20 demethylation (IEA) | GO:0035574
- obsolete induction by virus of host cell-cell fusion (IEA) | GO:0006948
- obsolete negative regulation of transcription during mitotic cell cycle (IEA) | GO:0007068
- obsolete reproduction (IEA) | GO:0000003
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- phenanthrene catabolic process via trans-9(R),10(R)-dihydrodiolphenanthrene (IEA) | GO:0018956
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of ATF6-mediated unfolded protein response (IEA) | GO:1903893
- positive regulation of axon guidance (IEA) | GO:1902669
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of macrophage chemotaxis (IEA) | GO:0010759
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of oligopeptide transport (IEA) | GO:2000878
- positive regulation of protein autoubiquitination (IEA) | GO:1902499
- positive regulation of siRNA processing (IEA) | GO:1903705
- positive regulation of tumor necrosis factor biosynthetic process (IEA) | GO:0042535
- primitive palate development (IEA) | GO:0062010
- programmed cell death (IEA) | GO:0012501
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- proton motive force-driven ATP synthesis (IEA) | GO:0015986
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of antisense RNA transcription (IEA) | GO:0060194
- regulation of beta-glucan metabolic process (IEA) | GO:0032950
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of chemorepellent activity (IMP) | GO:1903667
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of dendrite extension (IEA) | GO:1903859
- regulation of macrophage chemotaxis (IEA) | GO:0010758
- regulation of phosphatidylcholine biosynthetic process (IEA) | GO:2001245
- regulation of protein insertion into mitochondrial outer membrane (IEA) | GO:1903636
- regulation of response to type II interferon (IEA) | GO:0060330
- regulation of satellite cell activation involved in skeletal muscle regeneration (IEA) | GO:0014717
- release of sequestered calcium ion into cytosol by endoplasmic reticulum (IEA) | GO:1903514
- response to non-ionic osmotic stress (IEA) | GO:0010335
- response to osmotic stress (IEA) | GO:0006970
- response to reactive oxygen species (IEA) | GO:0000302
- sperm aster formation (IEA) | GO:0035044
- sterol metabolic process (IEA) | GO:0016125
- syncytium formation (IEA) | GO:0006949
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- tetrasaccharide transport (IEA) | GO:2001098
- xanthophyll biosynthetic process (IEA) | GO:0016123
- xanthophyll catabolic process (IEA) | GO:0016124
 Domains
Domains
- ( PF05462 ) Slime mold cyclic AMP receptor
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
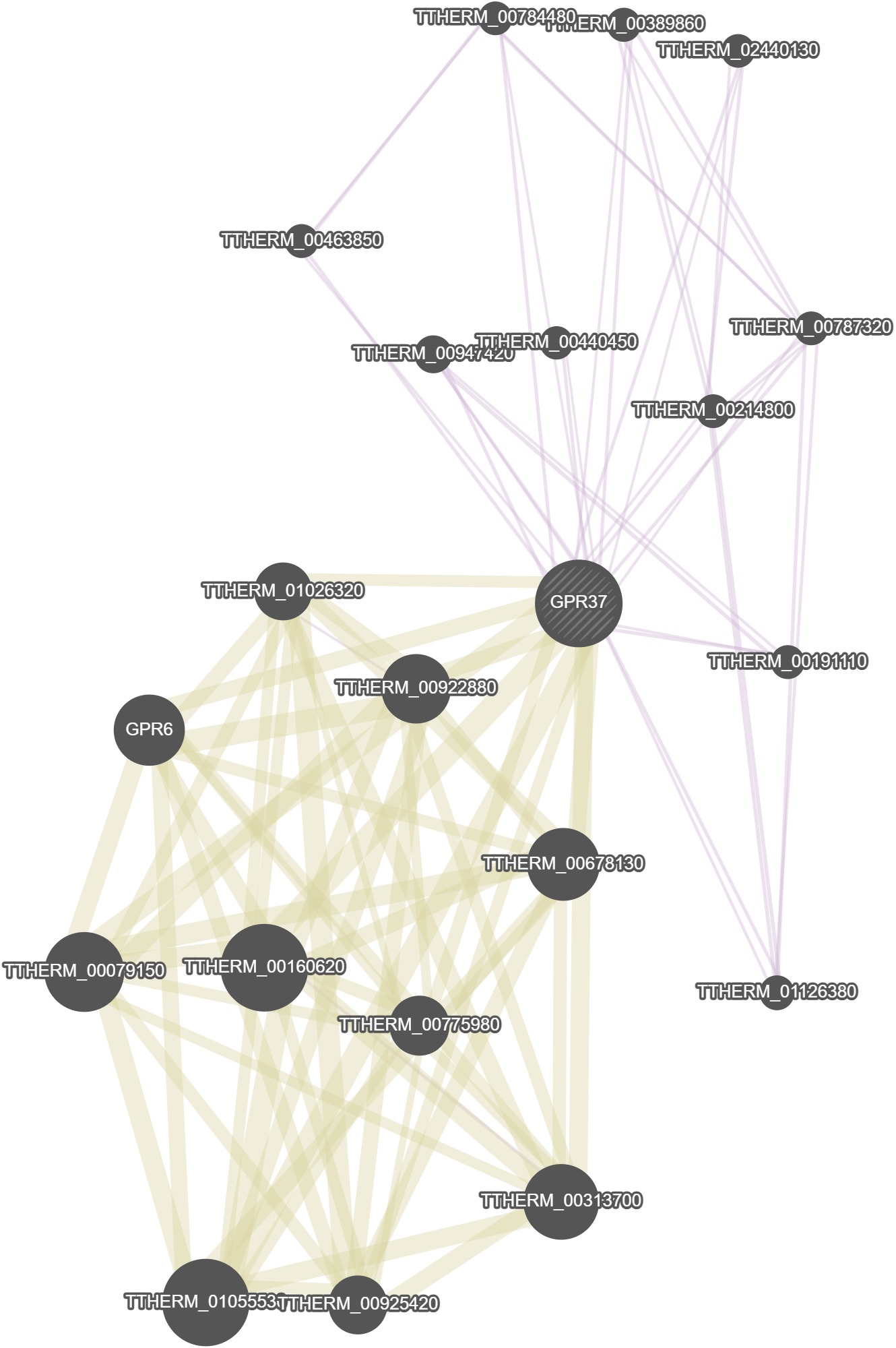
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
- ( SD01667 ) Macronucleus: GPCR3::neo3
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:28685161: Zou D, Hennessey TM (2017) Behavioral Effects of a Chemorepellent Receptor Knockout Mutation in Tetrahymena thermophila. mSphere 2(4):
 Sequences
Sequences
>TTHERM_00633170(coding)
ATGACGATTGTTTCTAACCACAATTAGGTCAACATTATATAGATTATAGCTATTGTTGTC
AGCTCTTTTTCTCTTCTAGGAGCTATATTTATTATTGGAATATTTTTGAGTTTCAAAAAG
CTCAGGGTTTTCCCCTTTATATATGTCTTTTGGCTCAGTGTAAGTGACTTAATATCTTCT
ATAGGGTACCTTCTTGTTACTGACTCGATTGGAGACACTCCTGAATAGCTTCAAGAGGCA
AATTTTACTTGTAAAACTTAGGCTTTAATGATTAATTTTGGATAGCTTGCCTCTATTTGT
TGGACTGCAGTTATTGCCTGGACTCTCTACAACTCCACCGTTAAAGTGGTAACAAACTTA
CAACAAAAAACAGAAACAAAAATGGTTGCGATTGCTTTTGGATTTCCCCTAATTATGTCA
TTGATACCTTTAATTTTAAATGAATATGGCCCATAAGGAGGCTGGTGCTGGATAAAAAGT
GAAGGCAGCAATAAAGTATTAACATTTATTTTGGTAGGAATAGAATTTTATATTCCTCTT
TGGATAGCATTCGTATTTAATGGCATAACAATATACAGAGTAATCAAATATGTCAAGAGT
ATAACTCAAGATAAAAATTAGCTGAAACTAATCAATAGACTAACACTATATCCTCTTATT
TTATTGGTTTGCTGGAGTGCTGGTACCGTAGAAAGAGTATTTGATTTCATCGGTATCTCG
TCAGAGATACTTTCATATTTGCACATCTTCTTTGGTGGATTGTAAGGATTTTTTAATGCT
TGTGTTTACGGTCTCAATTCAAATGTCCGTAAAGTTATCAGAGAAAATCTATGCTCAAGC
CTACCATGCTTTGCAAGTGCATATATATATGAAGAATAAAACCTTAATGAAGCTGAAGAT
CATCAGCAAGATAGTAAAAAAACTGCATCCATATCCTCTGGCGAAGGCCAAGTTGAAATG
GAACAACTTAATCAATGA>TTHERM_00633170(gene)
TCATTAAGAAGAATTTATTAGTAACTAGGCCAGAAAAAAAAAATTAAAAATTAATTAATT
AATTGATTGTTTATTTATTAAATTATTATTTAGCAAATACTTCTAAGACATATTATTGTT
ATTTATTTTTAGCGCACTTAGTTAGCTACAAATTATAAAGTAAAAAGCTTGAATTAAGCC
ATTAGCATAGAGTTATGACGATTGTTTCTAACCACAATTAGGTCAACATTATATAGATTA
TAGCTATTGTTGTCAGCTCTTTTTCTCTTCTAGGAGCTATATTTATTATTGGAATATTTT
TGAGTTTCAAAAAGCTCAGGGTTTTCCCCTTTATATATGTCTTTTGGCTCAGTGTAAGTG
ACTTAATATCTTCTATAGGGTACCTTCTTGTTACTGACTCGATTGGAGACACTCCTGAAT
AGCTTCAAGAGGCAAATTTTACTTGTAAAACTTAGGCTTTAATGATTAATTTTGGATAGC
TTGCCTCTATTTGTTGGACTGCAGTTATTGCCTGGACTCTCTACAACTCCACCGTTAAAG
TGGTAACAAACTTACAACAAAAAACAGAAACAAAAATGGTTGCGATTGCTTTTGGATTTC
CCCTAATTATGTCATTGATACCTTTAATTTTAAATGAATATGGCCCATAAGGAGGCTGGT
GCTGGATAAAAAGTGAAGGCAGCAATAAAGTATTAACATTTATTTTGGTAGGAATAGAAT
TTTATATTCCTCTTTGGATAGCATTCGTATTTAATGGCATAACAATATACAGAGTAATCA
AATATGTCAAGAGTATAACTCAAGATAAAAATTAGCTGAAACTAATCAATAGACTAACAC
TATATCCTCTTATTTTATTGGTTTGCTGGAGTGCTGGTACCGTAGAAAGAGTATTTGATT
TCATCGGTATCTCGTCAGAGATACTTTCATATTTGCACATCTTCTTTGGTGGATTGTAAG
GATTTTTTAATGCTTGTGTTTACGGTCTCAATTCAAATGTCCGTAAAGTTATCAGAGAAA
ATCTATGCTCAAGCCTACCATGCTTTGCAAGTGCATATATATATGAAGAATAAAACCTTA
ATGAAGCTGAAGATCATCAGCAAGATAGTAAAAAAACTGCATCCATATCCTCTGGCGAAG
GCCAAGTTGAAATGGAACAACTTAATCAATGAACAAAAAATATAAATCAAATCAATAAAT
TGAGATGAAATAAATATAAATATATTTAAATAAATTCTATTAACAAAAAAGAAATGGAAA
AGCTTTTTTAAGAAAGATGAAACAAACAAACAAAATAAAATAAAATAATAATATACATTT
GTTTATACAAATTAAATTATAATAGTTAATTAATGATTTATAAATAATTTAATATTATTT
TTACATTTAATATAATATTGGAATAATGATAACCATTTAAAAGAGATGTCTGACTTGAGA
GAAAGAAAATCAAACCAAATCAGTTATGAAATTTTATAAAGCAAATCATAGATTAACAAA
TAATGCCTTGCTAGCCTTTATCATAATACAAACTTATTTAATCGCTTTACACAAAAATAA
AAATTGCTTGAAGATTATGCATGTAAATCAGACAATTTATGTATCTTCATTCTTTACTTA
ATCTTCAATGAGGAGAACATCTTTTACTTTCATTTTTTAAGGTTCTTGGTCCTTTTAATC
CAAGCCAGCTATTTATAAAGTTTCTTTTTCTTTATATACAAAATTGCCTTCTAAGAACCC
CTACAGATCATCACTGGTATCGTAATTTCTGCTTATTGTTCAAAAATTACTTAATGGTTG
AACATAAATACATACATATATAAATAACTTAATAATAATTAGAAATAAATTCTTTGTGTT
AATTTACCTCATAGGTGAAAATGATGGCAGTCGTTGCTGCTACTACTATCCTTGTGCAGC
ATTTTGGTTAAAAATAGTTTAGCCTGTATTATCTTCTCCATTAATTTCTTTTAAATTTAT
GACAATATTAGCTTAATCTTCCTTTGTACTATTAATAATATTTATATTGTAATTTTTATT
ATCTATTTCCATTTTTAACAAGCAAAAA>TTHERM_00633170(protein)
MTIVSNHNQVNIIQIIAIVVSSFSLLGAIFIIGIFLSFKKLRVFPFIYVFWLSVSDLISS
IGYLLVTDSIGDTPEQLQEANFTCKTQALMINFGQLASICWTAVIAWTLYNSTVKVVTNL
QQKTETKMVAIAFGFPLIMSLIPLILNEYGPQGGWCWIKSEGSNKVLTFILVGIEFYIPL
WIAFVFNGITIYRVIKYVKSITQDKNQLKLINRLTLYPLILLVCWSAGTVERVFDFIGIS
SEILSYLHIFFGGLQGFFNACVYGLNSNVRKVIRENLCSSLPCFASAYIYEEQNLNEAED
HQQDSKKTASISSGEGQVEMEQLNQ